Dissertation on Growth and Chemical Production of Cyanobacteria
Info: 10222 words (41 pages) Dissertation
Published: 17th Nov 2021
Tagged: ChemistryEnvironmental Studies
Effect of Environmental Conditions on Growth and Ethanol Production in Cyanobacteria overexpressing BiBP
Abstract
The purpose of this study is to investigate the effects of light intensity and carbon dioxide concentration on growth and ethanol production of the model cyanobacteria Synechocystis PCC 6803 and Synechococcus PCC 7002 (hereafter referred to as 6803 and 7002, respectively). Cyanobacteria represents an ideal host for chemical production through metabolic engineering due to their ability to convert CO2 into useful compounds using energy from light. Models of C3 photosynthesis and metabolic control analyses have identified sedoheptulose-1,7-bisphosphatase (SBPase) in the CBB Cycle as a potential target of overexpression to increase carbon flux. This has been proven by numerous transgenic plant studies under a range of environmental conditions, as well as in cyanobacteria, which contains a bifunctional form of the enzyme, fructose-1,6-/sedoheptulose-1,7-bisphosphatase (BiBP).
Overexpressing BiBP in cyanobacteria could therefore, in theory, result in increased growth and chemical production. However, previous work from our lab has shown that overexpression of BiBP in 6803 strains producing ethanol does not lead to increased growth or ethanol production. In this work, the effect of environmental conditions on growth rate and ethanol production of the engineered 6803 and 7002 strains are examined. BiBP was overexpressed in 6803 and 7002 strains, as well as 6803 ethanol producing strains, and grown under saturating light intensity. Growth was analysed by measuring the optical density (OD) of the strains using the Multi-Cultivator 1000 (MC1000), and ethanol production was quantified using high-pressure liquid chromatography (HPLC). The amount of BiBP was quantified using Selective Reaction Monitoring Mass Spectrometry (SRM MS). None of the BiBP overexpressing strains showed an increase in growth relative to wild type, and BiBP overexpression did not result in an increase in ethanol production relative to the wild type.
Click to expand Contents and Abbreviations
Contents
- Introduction
- Metabolic Engineering
- BiBP
- Photosynthesis and the CBB Cycle
- Importance of SBPase/BiBP in controlling carbon flux through the CBB Cycle
- Effect of light intensity on carbon flux of BiBP-overexpressing strains
- Effect of [CO2] on carbon flux of BiBP-overexpressing strains
- Project aims
- Materials and Methods
- Culture Conditions
- Constructs
- eYFP Fluorescence
- Triparental Conjugation
- Growth Analysis
- Ethanol Measurements
- Sample Preparation for SRM Mass Spectrometry Analysis
- Protein Quantification by SRM
- Results
- Confirmation of pDF constructs expression – eYFP fluorescence
- Growth Analysis of BiBP-overexpressing 6803 and 7002
- Growth Analysis and Ethanol Production Measurements
- Discussion
- Next Steps
- References
Abbreviations
6803 – Synechocystis PCC 6803
7002 – Synechococcus PCC 7002
2PGA – 2 Phosphoglyceric acid
3PGA – 3 Phosphoglyceric acid
ADH – Alcohol Dehydrogenase
BiBP – Fructose-1,6-/Sedoheptulose-1,7-bisphosphatase
CBB Cycle – Calvin-Benson-Bassham Cycle
DHAP – Dihydroxyacetone phosphate
E4P – Erythrose-4-phosphate
FBP – Fructose-1,6-bisphosphate
FBPase – Fructose-1,6-bisphosphatase
F6P – Fructose-6-phosphate
G3P – Glyceraldehyde-3-phosphate
G6P – Glucose-6-phosphate
Gent – Gentamycin
HPLC – High-Pressure Liquid Chromatography
Jmax – Maximum rate of photosynthetic electron transport
MC1000 – Multi-Cultivator 1000
OAA – Oxaloacetate
OD – Optical Density
PDC – Pyruvate Decarboxylase
PEP – Phosphoenolpyruvate
RBS – Ribosomal Binding Site
Rubisco – Ribulose Bisphosphate Carboxylase/Oxygenase
RuBP – Ribulose Bisphosphate
Ru-5-P – Ribulose-5-phosphate
SBP – Sedoheptulose-1,7-bisphosphate
SBPase – Sedoheptulose-1,7-bisphosphatase
S7P – Sedoheptulose-7-phosphate
SRM – Selective Reaction Monitoring
TCA Cycle – Tricarboxylic Acid Cycle
μE – Microeinsteins
Vc,max – Maximum carboxylation rate of Rubisco
Introduction
1.1 Metabolic Engineering
Metabolic engineering aims to alter the metabolism of a host organism in order to produce desired chemical products not normally made by the host by introducing metabolic networks. To this end, cyanobacteria are an attractive host organism due to its ability to directly fix inorganic carbon from carbon dioxide into complex organic compounds using energy from light through photosynthesis [1,2]. However, introduction of non-native metabolic networks inevitably results in disturbances in the metabolic flux of the host due to interactions with the native metabolism. In order to maximize the efficiency of introduced metabolic networks in the host, tweaking of the native metabolism might provide a way in which the balance of the metabolic flux can be shifted in favour of the artificial network. Ethanol production represents a good model metabolic network due to its simplicity, with only 2 enzymes, Pyruvate Decarboxylase (PDC), which converts pyruvate into acetaldehyde, and Alcohol Dehydrogenase (ADH), which converts the acetaldehyde into ethanol, being introduced into the host organism. Ethanol is also a simple and non-toxic product, and can be readily detected and quantified using High Pressure Liquid Chromatography (HPLC). Ethanol production can therefore be used as a model to study the effects of altering the native metabolism of the host on the yield of the introduced metabolic network.
1.2 BiBP
Fructose-1,6-/sedoheptulose-1,7-bisphosphatase (hereafter referred to as BiBP), is a member of the FBPase family that exists exclusively in cyanobacteria. It is a bifunctional enzyme, catalysing simultaneously two reactions – the removal of a phosphate group each from fructose-1,6-bisphophate and sedoheptulose-1,7-bisphosphate to give fructose-6-phosphate and sedoheptulose-7-phosphate, respectively [4,5,6]. In plants, these two reactions are catalysed separately by the enzymes Fructose-1,6-bisphosphatase (FBPase) and Sedoheptulose-1,7-bisphosphatase (SBPase).
There are two types of FBPases in plants depending on the location of the enzyme: cytosolic FBPase is involved in the gluconeogenesis pathway and the synthesis of sucrose, while chloroplastic FBPase is involved in RuBP regeneration in the CBB Cycle and the synthesis of starch [4]. Both types of FBPases require divalent metal ions as cofactors, but cytosolic FBPase is regulated by AMP while chloroplastic FBPase is regulated by the light-dependent ferrodoxin-thioredoxin system [4]. BiBP also requires Mg2+ ions as cofactors, but it is different from both types of FBPases in that it is regulated by both the ferrodoxin-thioredoxin system as well as AMP [4].
1.3 Photosynthesis and the CBB Cycle
Photosynthesis can be divided into two main parts: the light-dependent reactions and the light-independent reactions, also known as the Calvin-Benson-Bassham Cycle (CBB Cycle). In the light-dependent reactions, energy from photons are used to split water, releasing excited electrons that are passed through the electron transport chain to generate reducing power in the form of NADPH, as well as ATP. NADPH and ATP are then fed into the CBB Cycle, where they are used in a series of reactions in which inorganic carbon in the form of carbon dioxide is fixed onto organic carbon compounds. These organic carbon compounds are then fed into various other metabolic pathways and serve as precursors for the synthesis of other metabolites.
The CBB Cycle itself can be thought of in three main parts: carbon fixation, carbon reduction, and RuBP regeneration. Carbon dioxide is fixed onto an acceptor molecule, Ribulose Bisphosphate (RuBP), a reaction catalysed by the enzyme Ribulose Bisphosphate Carboxylase Oxygenase (Rubisco), to generate 2 molecules of 3-phosphoglycerate, one of which is fed into other metabolic pathways while the other is phosphorylated by ATP and then reduced by NADPH to produce Glyceraldehyde-3-phosphate (G3P). G3P undergoes a series of reactions, consuming one ATP in the process, to regenerate the acceptor molecule RuBP, completing one turn of the cycle.
Among the series of reactions in the regeneration phase are the dephosphorylations of FBP and SBP, the reactions catalysed simultaneously by BiBP in cyanobacteria, and separately by FBPase and SBPase respectively in plants. The SBPase reaction is the first reaction that commits the carbon compound towards RuBP regeneration, and is therefore unique to the CBB cycle, while the product of the FBPase reaction, fructose-6-phosphate (F6P), can either be further processed towards RuBP regeneration or be converted to glucose-6-phosphate (G6P) and be fed into the sucrose and starch biosynthesis pathways [8, 11, 12, 20]. BiBP sits at the branch point of carbon partitioning between RuBP regeneration and carbohydrate synthesis, and is therefore a key enzyme in the CBB Cycle by driving the regeneration of RuBP [7].
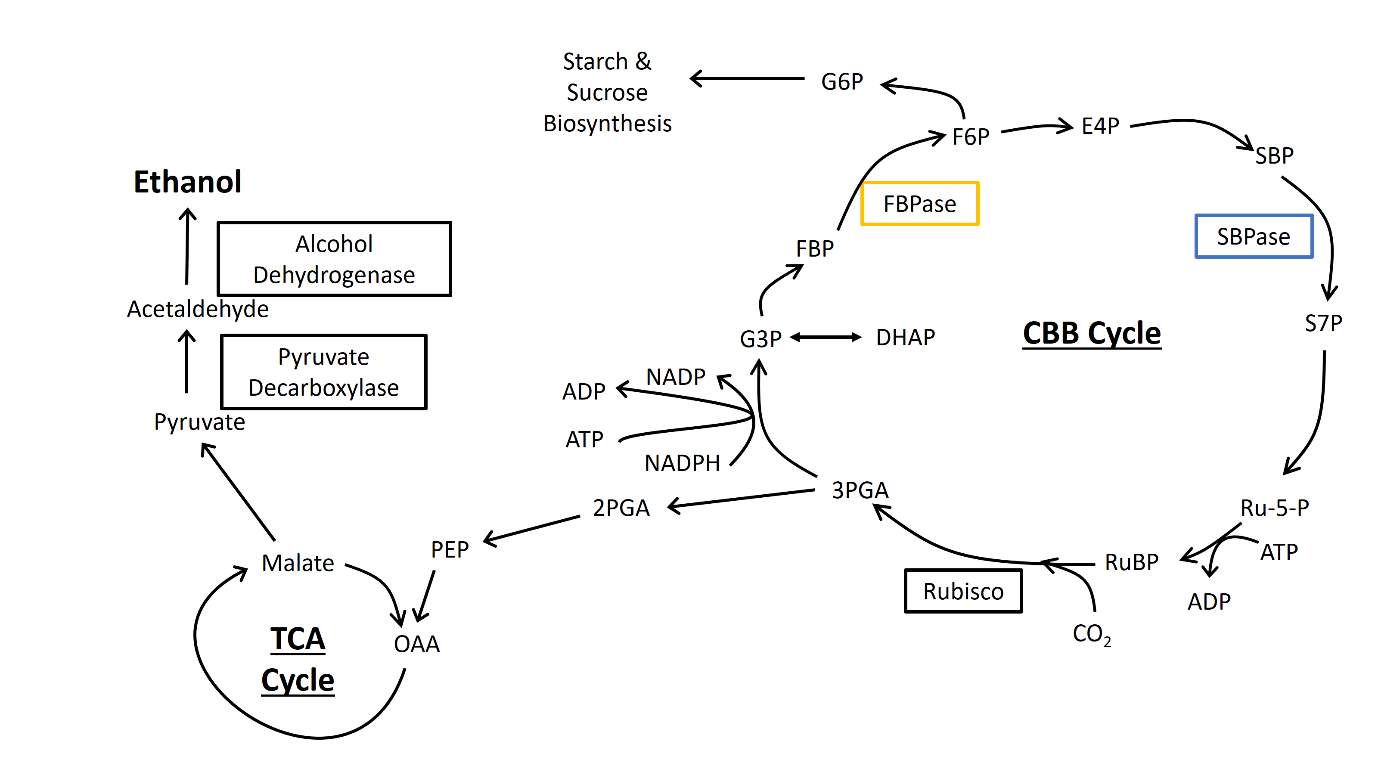
Fig.1 Schematic of photosynthetic carbon metabolism and the engineered ethanol production cassette.
1.4 Importance of SBPase/BiBP in controlling carbon flux through the CBB Cycle
Numerous theoretical models of C3 photosynthesis, as well as metabolic control analyses, have highlighted SBPase as a potential target for improving carbon flux through the CBB Cycle [7 – 12]. At atmospheric conditions, carbon flux through the CBB cycle was found to be limited by two phases of the cycle: the carboxylation of RuBP by Rubisco and regeneration of RuBP, and a balance is maintained at atmospheric conditions such that neither process exerts complete control on the photosynthetic rate [8, 9, 14, 19]. While numerous enzymes are involved in the regeneration of RuBP, many of which catalyse irreversible reactions, SBPase has been shown to exert the greatest control on the maximum RuBP regeneration capacity (Jmax), and that the carbon assimilation rate (A) is most sensitive to SBPase, as well as Rubisco [7]. It has also been shown theoretically that the relative amounts of different enzymes in the CBB cycle are not optimal for achieving the maximum photosynthetic rate, both under atmospheric and elevated [CO2] [7]. Specifically, the amounts of SBPase and aldolase, both of which operate in the RuBP regeneration phase of the CBB cycle, and Rubisco, were shown to be lower than necessary to achieve a higher photosynthetic rate, while enzymes in the photorespiratory pathway are well in abundance [7].
The importance of the SBPase reaction in RuBP regeneration rate and carbon assimilation rate highlighted in theoretical studies has been validated by experimental data. Antisense tobacco plants displaying a range of decreased SBPase activities relative to the wild type showed that Jmax is very sensitive to the level of SBPase activity, and that RuBP regeneration rate decreases linearly with small decreases in SBPase activity [19]. This sensitivity of RuBP regeneration rate towards SBPase activity shows that the wild type level of SBPase activity is limiting to the rate of RuBP regeneration, and that normal levels of SBPase in wild type plants may be only sufficient to maintain the wild type rate of RuBP regeneration [19].
This impact on the regeneration phase of the CBB cycle is reflected in reduced carbon assimilation rates and growth rates compared with wild type, as well as a drastic reduction in the levels of starch in antisense tobacco plants, with a decrease to 5% wild type starch levels seen in transgenic strains displaying less than 15% of wild type SBPase activity [8]. This altered carbon partitioning is in keeping with the role of SBPase as controlling the balance between carbon export into starch or sucrose biosynthesis and RuBP regeneration. In agreement with these observations, increase in the SBPase reaction by overexpressing either BiBP or SBPase in transgenic plants has been shown to increase photosynthetic rates and growth. Overexpression of BiBP from the cyanobacteria Synechococcus PCC 7942 (hereafter referred to as 7942) in tobacco grown in atmospheric conditions resulted in a 1.5-fold increase in final dry matter and a 1.24-fold increase in carbon assimilation rate [20].
The same group then attempted to clarify the relative contributions of FBPase and SBPase to this increase in photosynthetic rate by overexpressing each enzyme independently in transgenic plants, showing that while overexpressing FBPase alone resulted in the increased photosynthetic rate, a much higher increase in its activity was required to achieve the phenotype compared to that of SBPase, highlighting SBPase as the most important control point in RuBP regeneration with FBPase being a secondary control point [21]. This is backed up by antisense studies of SBPase and FBPase, which showed that a detrimental phenotype was only observed when FBPase activity was reduced to less than 14% of the wild type, while reducing SBPase activity to 71% of the wild type resulted in a marked reduction in photosynthetic activity [21].
Overexpressing SBPase in tobacco grown in ambient [CO2] by Lefebvre et al. also showed increased photosynthetic rates, as well as increased accumulation of sucrose and starch during the day, relative to the wild type control [15]. Increase in biomass and leaf area relative to the wild type were also observed [15]. This result was replicated by Rosenthal et al. under the same ambient [CO2] condition [17].
Studies with other transgenic plants have shown similar results. Overexpression of SBPase in wheat has also shown increased carbon assimilation rates under light saturating conditions, resulting in increased plant biomass, seed weight, and number of ears per plant compared with the wild type [14]. Transplastomic lettuce overexpressing BiBP from 7942 were shown to have a 1.3-fold increase in photosynthetic capacity relative to the wild type, as well as a 1.6-fold increase in fresh weight, larger leaf sizes, and increased number of leaves compared with wild type [24].
Tomatoes overexpressing SBPase showed not only increased photosynthetic rates, sucrose and starch levels, leaf area, and biomass accumulation, but also an increased tolerance to chilling stress [13]. Similarly, in rice overexpressing SBPase, tolerance to salt and heat stress was observed; however, there was no increase in growth compared to wild type [25]. Interestingly, another study overexpressing BiBP rather than SBPase in rice were able to show increases in leaf dry mass per area, plant height, as well as soluble carbohydrate content relative to wild type [26]. It is possible that increased activities of both SBPase and FBPase are required for increased photosynthetic capacity and growth rate, and the exact amount of increased activity may vary from species to species.
BiBP overexpression in cyanobacteria has shown similar results in terms of growth. Liang et al. showed that overexpression of BiBP placed under the strong, light-driven psbA2 promoter in Synechocystis PCC 6803 resulted in increased growth rate and biomass accumulation, as well as increased chlorophyll a content, relative to the wild type [27]. Unpublished data from De Porcellinis et al. using the marine cyanobacterium Synechococcus PCC 7002 showed that other than increased growth rate in terms of doubling time, cell sizes of the transgenic lines were also much larger when measured. The rate of photosynthetic electron transport, represented by O2 evolution, was also increased.
This study also validated antisense plant experiments reporting an increase in soluble carbohydrate content while also highlighting the fact that storage carbohydrate, in the form of glycogen, was decreased, indicating that there is an altered partitioning of carbon end products in the CBB cycle. This is of interest in the context of metabolic engineering towards chemical production, since overexpression of BiBP could lead to increased carbon flux towards excreted chemicals such as ethanol.
This potential impact of BiBP overexpression on chemical production was evaluated in a study by Ogawa et al. using Euglena gracilis, a photosynthetic, single-celled eukaryote. E. gracilis produces paramylon when grown under aerobic conditions, and when switched to anaerobic conditions, converts the paramylon to wax esters, a valuable chemical. Not only was biomass production significantly increased in transgenic lines, paramylon accumulation and subsequent wax ester production was also drastically elevated relative to the wild type [23]. This proved that overexpressing BiBP not only results in increased growth and biomass accumulation, but it also has the potential to increase chemical production in model organisms through elevated carbon flux through the CBB Cycle.
With these observations in mind, previous work from our lab sought to investigate the effect of BiBP overexpression on the growth rate and ethanol production of engineered Synechocystis PCC 6803. An ethanol production cassette with the genes pdc (pyruvate decarboxylase) and adh (alcohol dehydrogenase), followed by bibp, were integrated into the neutral site slr0168 and placed under the control of the lac-inducible PA1lacO1 promoter. However, overexpression of BiBP did not result in any change in growth rate or ethanol production compared to control strains.
A possible reason for this unexpected result is the conditions under which the strains where cultured in. The cultures were grown under 60 μE light intensity, with 1% CO2 at 30˚C; while SBPase has been shown to be an important regulator of carbon flux in the CBB Cycle in higher plants and cyanobacteria, overexpressing SBPase or BiBP may only provide a positive impact on photosynthetic rate and growth when RuBP regeneration is limiting, which is not the case under all environmental conditions [11].
1.5 Effect of light intensity on carbon flux of BiBP-overexpressing strains
Since the CBB cycle utilizes the products of the photosynthetic electron transport chain for carbon reduction, it naturally follows that the light-independent reactions are limited by the light-dependent reactions, which in turn are limited by the photosynthetic photon flux density (PPFD). Theoretical models postulating the limiting effects of SBPase activity all assume that the CBB cycle is not limited by the amount of NADPH and ATP; therefore, any benefit in increasing SBPase activity would only be seen under light saturating conditions where NADPH and ATP are not limiting carbon reduction in the CBB cycle [7, 9, 10]. This is backed by experimental evidence both in higher plants and cyanobacteria.
Lefebvre et al. demonstrated that transgenic tobacco overexpressing SBPase did not show any increases in growth or photosynthetic rate when grown during the winter with shorter days and lower light intensity, in contrast to the phenotype displayed when grown under high light intensity [15]. This is further quantified by Miyagawa et al., who showed that significant increases in photosynthetic activity in tobacco overexpressing BiBP were only observed above a threshold of light intensity, with the highest increase in photosynthetic activities over the wild type achieved when transgenic plants were grown under fully light saturating conditions; the same authors found the same threshold when overexpressing SBPase or FBPase alone in tobacco [20, 21].
Diurnal measurements of transgenic tobacco also showed that carbon assimilation was most elevated relative to the wild type at midday, while at the beginning or end of the day there was no increase [17]. Unpublished data from De Porcellinis et al. showed that in 7002 overexpressing BiBP, while increase in the photosynthetic and growth rates were observed under both high light and normal light conditions, the phenotypes were more pronounced in strains cultured under light saturation. Liang et al. showed in 6803 that the positive phenotype associated with BiBP overexpression was only observed in strains cultured under 100 μE light intensity, while those cultured under 15 μE light intensity did not display the same phenotype [27].
Interestingly, strains overexpressing BiBP actually displayed slower growth rates than the wild type and a slower increase in chlorophyll a content [27]. This proves that while under high light intensity, overexpression of BiBP results in faster growth, under low light intensity, overexpression of BiBP actually inhibits growth of 6803 cells. It is noteworthy that this trend is present not only in the overexpression of BiBP, but in overexpression of Rubisco, FBA, and TK as well [27].
Evidently, when light is limiting, attempts at speeding up the CBB Cycle is counterproductive; since the light-dependent reactions are actually the rate-limiting steps rather than the CBB Cycle. Specifically, since RuBP regeneration requires NADPH and ATP, it is therefore driven by the photosynthetic electron transport and thus dependent on light intensity; RuBP regeneration may only be limiting under light saturation.
1.6 Effect of [CO2] on carbon flux of BiBP-overexpressing strains
Another growth condition that may affect the efficacy of BiBP overexpression is the concentration of CO2. Since the CBB cycle is co-limited by the regenerative phase and the carbon fixation reaction at atmospheric conditions, shifting the balance between these two processes depends on the relative activities of the two main enzymes driving each process, namely SBPase and Rubisco, respectively [8, 9]. Rubisco is a major limiting factor at atmospheric conditions because of its low specificity, resulting in a low turnover rate.
Other than the carboxylation of RuBP, Rubisco also catalyses the oxygenation of RuBP due to its inability to distinguish CO2 and O2, resulting in a useless side reaction whose product is toxic and places a metabolic burden on the organism when converting it into a non-toxic compound to be reused in the CBB cycle. At atmospheric CO2, the equilibrium between the carboxylation and oxygenation reactions is such that the carboxylation of RuBP is the major limiting factor in carbon flux [8, 9, 10, 14, 17].
At elevated concentrations of CO2, however, the equilibrium shifts in favour of the carboxylation reaction, reducing the oxygenation reaction to a negligible rate [11]. Rubisco is no longer limiting under elevated [CO2], and the limitation on the CBB cycle shifts to RuBP regeneration [14, 17]. Theoretically, increasing RuBP regeneration capacity by elevating the SBPase reaction could therefore be the most useful under saturated [CO2].
The hypothesis that overexpressing SBPase or BiBP under elevated [CO2] would result in the most productive impact on photosynthesis is validated by experimental data. Transgenic tobacco overexpressing BiBP displayed the highest increase in photosynthetic activity over the wild type under saturated CO2 conditions [17].
While transgenic tobacco also showed increased biomass relative to the wild type under ambient [CO2], this increase was more pronounced when grown under elevated [CO2], as well as displaying higher accumulations of soluble carbohydrates [20]. These results were also validated in wheat grown under saturating [CO2] [14]. Euglena gracilis grown under both high light intensity and [CO2] displayed both increased cell volume and growth rate, while strains grown under high light only showed only increased cell volume, and grown under ambient conditions showed no increase in growth rate above the wild type [23].
In fact, growth of the wild type was inhibited under ambient [CO2] at high light due to the fact that excess reductant produced by the electron transport chain were not able to be used up by the CBB cycle; this was confirmed by the rescue of the wild type growth rate by increasing [CO2] [23].
On the other hand, high [CO2] has not always been shown to be required for increased growth and photosynthetic rate in SBPase- or BiBP-overexpressing strains. Soybean overexpressing BiBP showed increased carbon assimilation and photosynthetic rate under both ambient and elevated [CO2] compared to the wild type, while transgenic strains were able to maintain seed yield under elevated [CO2] and temperature and the wild type was not [22].
Transgenic tobacco and wheat were shown to have increased photosynthesis under both ambient and elevated [CO2], even though the increases were more pronounced under elevated [CO2], and 6803 strains overexpressing BiBP were shown to have higher growth rates at ambient [CO2] under high light intensity [14, 20, 21, 27]. Therefore, while elevated [CO2] theoretically maximizes the benefits of SBPase or BiBP overexpression by increasing the carboxylation reaction of Rubisco and shifting the rate limiting step to RuBP regeneration, experimental data shows that this is not necessarily a requirement for increased carbon flux through the CBB cycle, even though it could accentuate the positive effect of BiBP on photosynthesis.
1.7 Project aims
To test the hypothesis that previous work in our lab showing that BiBP overexpression resulted in no increase in growth rate or ethanol production in 6803 ethanol producing strains because of suboptimal light intensity and [CO2], BiBP from 6803 was overexpressed in wild type 6803 and 7002 and cultured under high light intensity for 7 days, with the goal of replicating the results shown by Liang et al. and de Porcellinis et al. in terms of growth enhancement. 6803 ethanol-producing strains overexpressing BiBP were re-evaluated under higher light intensity, and growth rate and ethanol production measured after 7 days.
Materials and Methods
2.1 Culture conditions
6803 strains were cultured in BG-11 media. Cultures were grown in flasks containing 25mL of BG-11 media at 30˚C, 130 rotary shaking, at ambient [CO2] and light intensity, and supplemented with spectinomycin at a final concentration of 50 μg/mL or gentamycin at a final concentration of either 50 μg/mL on agar plates or 0.5 μg/mL in liquid media. Growth was monitored at OD730nm with the use of Infinite 200 Pro spectrophotometer (Tecan Group Ltd. Switzerland).
7002 strains were cultured in A+ media supplemented with vitamin B12 at a final concentration of 4 μg/mL. Cultures were grown in flasks containing 25mL of A+ media at 30˚C, 130 rotary shaking, at ambient [CO2] and light intensity, and supplemented with spectinomycin at a final concentration of 50 μg/mL or gentamycin at a final concentration of 50 μg/mL. Growth was monitored at OD730nm with the use of Infinite 200 Pro spectrophotometer (Tecan Group Ltd. Switzerland).
E. coli strains were grown at 37˚C in Luria-Broth medium supplemented with the appropriate antibiotic at the following final concentrations: spectinomycin at 50 μg/mL, gentamycin at 10 μg/mL, chloramphenicol at 50 μg/mL, and carbenicillin at 50 μg/mL.
2.2 Constructs
Two constructs made by previous members of the lab derived from the RSF1010 self-replicating vector harbouring the bibp gene from 6803, pDF-Gent_lac_BiBP and pDF-Gent_pc143_BiBP, were used in the growth analysis experiments. An RSF1010-derived plasmid containing the eYFP gene was used as a control for the constructs. The constructs were extracted from E. coli DH5α glycerol stocks using a Qiagen Extraction Mini-prep kit, the concentration of DNA was quantified using the Infinite 200 Nanoquant by Tecan, and samples were sent to Source Biosource for diagnostic sequencing to confirm the presence of the bibp gene.
2.3 eYFP Fluorescence
pDF-eYFP, an RSF1010-derived plasmid containing the gene for eYFP, was used as a control for the expression of the pDF constructs. After 7 days post IPTG induction, 100 μL of culture was added to a 96-well plate and absorption and fluorescence readings were taken using the Infinite 200 Pro spectrophotometer (absorption: 730 nm; excitation 503 nm; emission 540 nm). The fluorescence was normalized against optical density at 730nm.
2.4 Triparental Conjugation
E. coli cargo strain IY134 harbouring pRL623 were inoculated in 5mL LB supplemented with 50 μg/mL chloramphenicol and incubated overnight at 37˚C. pDF-Gent_lac_BiBP or pDF-Gent_pc143_BiBP plasmids, were transformed into competent IY134 cells by heat shock and the transformed cargo strain cells were inoculated into 5mL LB supplemented with 50 μg/mL chloramphenicol and incubated overnight at 37˚C. 1 mL of the overnight cultures were then spun down, washed 3 times with LB medium to remove the antibiotics, and resuspended in 500 μL LB.
E. coli conjugate strain IY135 containing pRL443 was inoculated in 5mL LB media supplemented with 50 μg/mL carbenicillin overnight at 37˚C. 1 mL of the cultures were then spun down, washed with LB medium 3 times to remove the antibiotics, and resuspended in 500 μL LB.
6803 or 7002 strains were cultured to OD730nm of 0.8-1.0. 1 mL of the cultures were then spun down and washed with BG-11 or A+ media twice and resuspended in 500 μL BG-11 or A+ media.
100 μL each of cargo strain, conjugate strain, and cyanobacteria were mixed and incubated for 2 hours at 60 μE under 1% [CO2] with 130 rotary shaking. The mixtures were then transferred to agar plates with no antibiotics and incubated for 2 days, after which 500 μL of BG-11 or A+ media were used to transfer the cells to an agar plate supplemented with the appropriate antibiotic. The plates were then incubated for 10-14 days.
2.5 Growth Analysis
Growth analysis was carried out in the Multi-Cultivator 1000 (MC1000). Starter cultures were grown at 30˚C under ambient light and [CO2] until they reached OD730nm values of at least 0.4. The cultures were then inoculated into 50 mL of BG-11 or A+, supplemented with the appropriate antibiotic and vitamin B12 in the case of A+, to an initial OD730nm of 0.2-0.3. IPTG was added to a final concentration of 1mM. All strains were cultured in the MC1000 for 7 days, with 6803 strains under 250 μE light intensity and 7002 strains under 1000 μE light intensity. OD720nm measurements were taken automatically every 30 minutes.
2.6 Ethanol Production
Existing ethanol-producing 6803 strains contained an ethanol production cassette of pdc and adh, as well as bibp, under the PA1LacO1 promoter integrated into the neutral site slr0168. After 7 days post-IPTG induction, 1 mL of each culture was spun down at 4000g for 10 minutes. The supernatant was then analysed. Ethanol was quantified using the 1200 series HPLC system from Agilent Technologies. The eluent was 5 mM sulfuric acid in HPLC grade water with a flow rate of 0.6 mL/min. A refractive index detector was used to detect the signal.
2.7 Sample preparation for SRM Mass spectrometry analysis
After 7 days post-IPTG induction, 25 mL of each culture was spun down at 5000 rpm for 10 minutes at 4˚C. The pellets were resuspended in 1.5 mL BG-11 or A+ media and spun down again. The pellets were then resuspended in 500 μL Extraction buffer (1M Tris-HCL pH 8.0, 0.25 M EDTA, 1 M DTT) and spun down at 18,000 g at 4˚C for 10 minutes. The pellets were resuspended in 1000 μL Extraction buffer and 500 μL acid-washed 0.1mm silica/zirconia beads (nr. Sigma) were added. The cells were broken by vortexing for 7.5 minutes at 30 Hz. The supernatant was then spun down at 21,000g for 10 minutes at 4˚C to remove unbroken cells and debris.
2.8 Protein Quantification by SRM
Identification and quantification of BiBP was done using an established protocol developed by a previous member of the group. 3 peptides were used for BiBP and 2 peptides for AtpB, with 2-4 transitions per peptide. For 6803, relative quantification of BiBP was carried out using relative peak intensities normalized to the relative peak intensities of the AtpB native standard. Due to time constraints, an internal standard for 7002 with signature peptides and optimal transitions for MS detection was not available. Therefore, quantification of 6803 BiBP in 7002 strains was done without normalization.
The MS was configured with an Ion Drive Turbo V source, with Gas 1 and 2 set to 40 and 60 respectively, the source temperature set to 500˚C, and the ion spray voltage set to 5500 V. The MS, configured with high mass enabled, was used in ‘Triple Quadruple’ mode for Multiple Reaction Monitoring. Data was acquired and analysed using SCIEX software Analyst 1.6.1 and MultiQuant 3.0.
Results
3.1 Confirmation of pDF construct expression – eYFP fluorescence
B
A
C
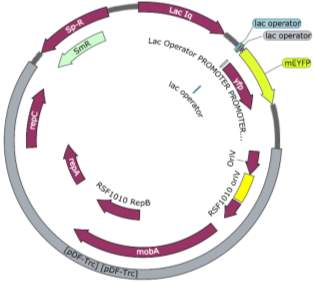
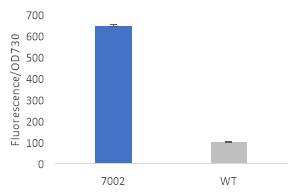
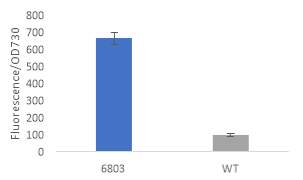
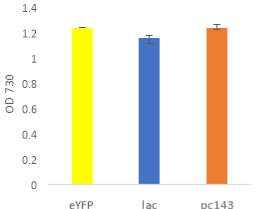
Fig.2 Fluorescence intensity normalized against cell density of eYFP construct in (A) 6803 and (B) 7002, compared to the respective wild type control. (C) Plasmid map of pDF-eYFP used as a control for RSF1010.
An eYFP construct consisting of the RSF1010 self-replicating vector and the eYFP gene under the Trc promoter was used as a control for the transformability and expression of the RSF1010 self-replicating vector in 6803 and 7002 (Fig. 2A). Cell density-normalised fluorescence confirmed that the RSF1010 self-replicating vector was transformed successfully by triparental conjugation in both 6803 and 7002 strains and expressed successfully by 1 mM IPTG induction (Fig. 2A, B).
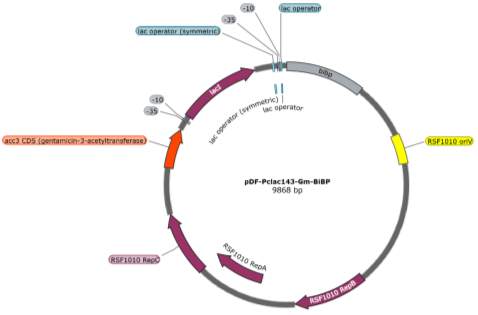
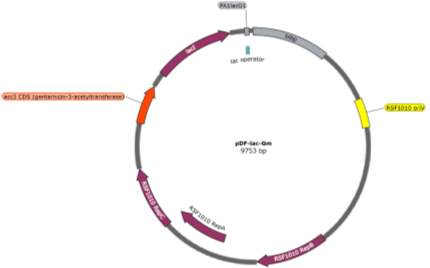
The two pDF-BiBP constructs were made by a previous member of the lab (Fig. 3). Both are derived from the RSF 1010 self-replicating vector and contain the gentamycin resistance gene acc3. Both constructs contain a lac-inducible promoter; in the case of the pDF-Gent_lac_BiBP, bibp was placed under the control of the PA1lacO1 promoter, a Lac promoter variant containing two natural lac operators placed upstream and downstream of the -10 sequence to facilitate DNA-looping and therefore tighter repression that has been confirmed to be effective in 6803 [28, 29, 30], while bibp in pDF-Gent_pc143_BiBP was placed under the control of the PcLac143 promoter developed by Markley et al. for use in 7002 [31].
Fig. 3 Plasmid maps of the pDF-BiBP constructs. (A) pDF-Gent_lac_BiBP (B) pDF-Gent_pc143_BiBP
3.2 Growth Analysis of BiBP-overexpressing 6803 and 7002
Av
Bv
Cv
Dv
Ev
Fv
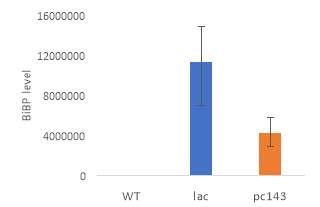
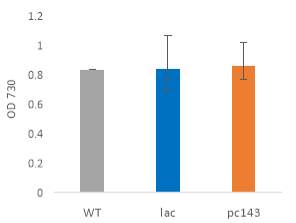
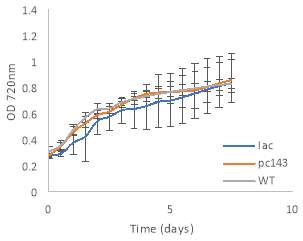
Fig.4 OD720nm of transgenic (A) 6803 strains under 250 μE light intensity and (B) 7002 strains under 1000 μE light intensity over 7 days. Final OD of (C) 6803 strains and (D) 7002 strains after 7 days. 6803 bibp expression levels of (E) 6803 strains normalized to AtpB native standard and (F) 7002 strains without normalization. 3 replicates were used for pDF-BiBP strains, while only one sample was used for the control strains.
After 7 days of culturing the strains under high light intensity, with 250 μE for 6803 strains and 1000 μE for 7002 strains, BiBP-overexpressing strains showed no increase in growth rate over control strains, represented by OD720 (Fig. 4A, B), despite successful bibp overexpression in 6803 and 7002, shown by SRM results (Fig. 4E, F). Final cell density of the cultures showed no increase in BiBP-overexpressing strains compared with control strains (Fig. 4C, D). 7002 strains showed visible signs of photobleaching after 3 days of culturing under 1000 μE light intensity, with the cultures turning yellow as opposed to their characteristic green colour. Although optical density continued to increase, growth rate was much slower than shown by Synechococcus 6803 strains, and stationary phase was never reached in the 7 days by 7002 strains (Fig. 4A, B).
3.3 Growth Analysis and Ethanol Production Measurements
Bv
Av
Cv
Dv
**
**
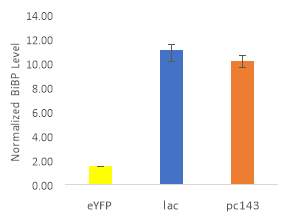
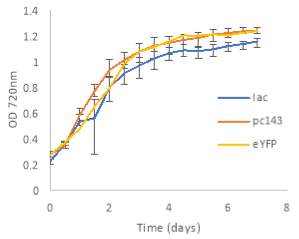
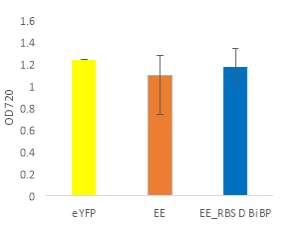
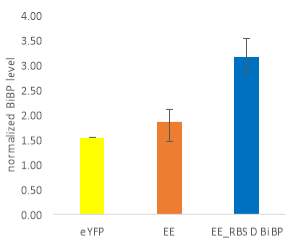
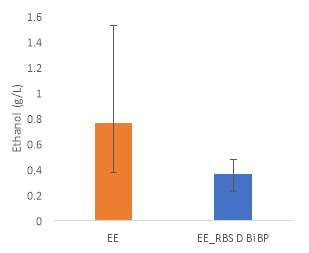
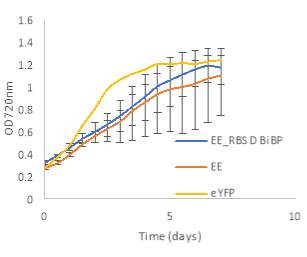
Fig.5 (A) OD720 of BiBP-overexpressing ethanol producers (EE_RBS D BiBP; blue), ethanol producing control (EE; orange), and eYFP control (yellow) over 7 days at 250 μE light intensity. (B) Final OD720 of the ethanol producing and control strains after 7 days. (C) Ethanol production of control and BiBP-overexpressing strain after 7 days. (D) bibp expression levels normalized to AtpB native standard. (**P<0.01)
Existing ethanol-producing 6803 strains over-expressing BiBP were re-evaluated under higher light intensity. Previous work involved culturing at 60 μE light intensity, while light intensity was increased to 250 μE light intensity in this work. Synechococcus 6803 strains transformed with the pDF-eYFP construct was used as a control for growth rather than the wild type due to a technical error. Growth rate of ethanol producers, regardless of BiBP overexpression, were slower than the control, shown by the later approach towards stationary phase (Fig. 5A), and there was no statistical difference in final OD720 after 7 days between the BiBP-overexpressing ethanol producer and the ethanol control and eYFP control strains (Fig. 5B).
Ethanol production of EE_RBS D BiBP was compared with an ethanol-producing control, EE, after 7 days in the MC1000 under 250 μE. While the average amount of ethanol produced by the BiBP-overexpressing strain was significantly lower than that of the control, when accounted for the outlier in one of the control replicates, there was no significant difference in the amount of ethanol produced between the control and BiBP-overexpressing strain (Fig. 5C). The amount of BiBP was, however, significantly higher in EE_RBS D BiBP relative to EE.
4 Discussion
SBPase has been demonstrated theoretically and experimentally to be an important control point for carbon flux through the CBB cycle. Previous work from our lab showed that BiBP-overexpression in ethanol-producing 6803 resulted in no increase in either growth or ethanol production. To understand why BiBP-overexpression did not lead to the increased photosynthetic carbon assimilation proven by experimental data by other groups, the effect of light intensity on the effect of BiBP-overexpression was examined. Liang et al. showed that 6803 overexpressing BiBP only show increased growth rate over the wild type under 100 μE light intensity while under 15 μE light intensity, transgenic strains actually grew slower than the wild type [27].
Unpublished results from de Porcellinis et al. showed that for 7002, high light intensity of 1500 μE results in a more pronounced increase in growth rate relative to the wild type, as well as increases in cell size, compared to light intensity of 250 μE. To try and replicate these results, wild type 6803 and 7002 strains were transformed with a self-replicating plasmid containing the BiBP gene, and growth was monitored over 7 days under 250 μE light intensity for 6803 strains and 1000 μE light intensity for 7002 strains. An existing ethanol producing 6803 strain overexpressing BiBP from our lab was grown under 250 μE for 7 days, and growth was monitored, and ethanol production was quantified after the 7 days.
None of the BiBP-overexpressing cyanobacterial strains exhibited any increase in growth under high light intensity relative to control strains, nor did the BiBP-overexpressing ethanol producer show any increase in ethanol production, in agreement with results of previous work from our lab, despite SRM results showing that bibp expression levels were higher in 6803 strains than the control strains. SRM results for 7002 were not available because an internal standard with optimal peptides and transitions had not yet been determined.
Theoretical and experimental data proved that BiBP-overexpression only has a positive impact on carbon assimilation under conditions where RuBP regeneration is not limited by ATP and NADPH [9, 15, 20, 21]. So why did culturing strains under light saturation, when ATP and NADPH should not be limiting, not result in any increase in growth when BiBP is overexpressed? The first reason may be that the cultures were not [CO2] saturated. BiBP is the most important enzyme in determining RuBP regeneration rate. Therefore, BiBP exerts greatest control on carbon assimilation when RuBP regeneration is the rate limiting step in the CBB cycle.
At atmospheric [CO2], RuBP regeneration and RuBP carboxylation co-limit photosynthetic rate [8, 9, 12]; since RuBP carboxylation is limited by the oxygenation reaction of Rubisco at atmospheric [CO2], increasing RuBP regeneration under these conditions still leaves the CBB cycle limited by the RuBP carboxylation rate. By elevating [CO2], the equilibrium between the carboxylation and oxygenation reactions of Rubisco is shifted such that the rate of RuBP oxygenation becomes negligible, improving the carboxylation rate and shifting the limitation of carbon flux towards RuBP regeneration [12].
Since the experiments were carried out at atmospheric [CO2], the lack of increase in carbon assimilation in BiBP-overexpressing strains may be due to RuBP regeneration not limiting the CBB cycle; hence, overexpressing BiBP results in an excess of RuBP that is not able to be consumed at the same rate by Rubisco.
On the other hand, Liang et al. showed an increase in growth rate in 6803 overexpressing BiBP under 100 μE light intensity at ‘ambient’ [CO2] [27]. How is that possible under conditions where limitation of carbon flux is still equally distributed between RuBP regeneration and carboxylation? The effect of different promoters controlling the bibp gene may be the answer. In this work, two constructs were used to overexpress BiBP, both based on the RSF1010 self-replicating vector and containing lac-inducible promoters PA1lacO1 and PcLac143. Liang et al. used the pPMQAK1 plasmid, also derived from the RSF1010 self-replicating vector, and placed bibp under the psbA2 promoter, a strong, light-induced promoter native to 6803 that drives transcription of photosystem genes [27].
While the amount of protein expressed in the transgenic lines was not mentioned in the work of Liang et al., it could be that the increase in growth rate of the transgenic strains under high light intensity as opposed to low light intensity was due to different levels of induction and transcription rates of bibp.
Numerous studies have made observations about a pleiotropic effect of BiBP or SBPase on Rubisco. Soybean overexpressing BiBP was shown to have an increase in the maximum carboxylation rate of Rubisco (Vc,max) [22]. In tobacco overexpressing SBPase, Vc,max was also increased relative to the wild type when grown in greenhouse conditions but not under open field conditions [15, 17].
Under atmospheric conditions, while total Rubisco activity remained the same, transgenic tobacco were shown to have increased initial activity of Rubisco relative to the wild type, corresponding to an increase in Rubisco activation in vivo [20, 21]. Unpublished data from de Porcellinis et al. also showed that under 3% [CO2] conditions, not only was total Rubisco activity elevated relative to the wild type, changes in the activities of other enzymes were observed. Specifically, Aldolase, involved in RuBP regeneration upstream of BiBP, was also increased in activity, while enzymes involved in carbon catabolism pathways such as Oxidative Pentose Phosphate Pathway and Glycolysis had either a decrease in abundance or alteration in post-translational modifications such that carbon catabolism was suppressed.
Since Rubisco activity, alongside RuBP regeneration, is limiting in the CBB cycle under atmospheric [CO2], the fact that overexpression of BiBP can result in a concomitant increase in Rubisco activity might explain why BiBP overexpression alone can result in increased photosynthetic rates and subsequent growth rate. De Porcellinis et al. also reported that the overexpression of Rubisco alone does not result in any of these changes.
Given that increase total Rubisco activity is not observed in all studies and under different conditions, does there exist a threshold in either light intensity or [CO2] or BiBP amount that results in an increase in Rubisco activity in BiBP-overexpressed strains? It is plausible that a sufficient amount of BiBP or SBPase might increase Rubisco activity to a level such that even at atmospheric [CO2], the rate of the carboxylation reaction is increased sufficiently to result in a noticeable increase in carbon assimilation. While it has been proposed that the increased RuBP content brought about by BiBP overexpression results in an increase in Rubisco activation, the exact mechanism by which BiBP affects Rubisco activation state and activity remains unknown.
Next Steps
Since the MC1000 only allows eight cultures at a time, culturing three replicates for each BiBP-overexpressing strain resulted in only one replicate used for each growth control strain; thus, statistical significance could not be determined for growth analysis data. Observations made in this work are, therefore, only preliminary, and any concrete interpretations can only be made once additional experiments are conducted for multiple replicates of the control strains.
The bibp was overexpressed in 6803 ethanol producers used in this study through a chromosomal integration cassette with bibp as the third gene of the operon, downstream of pdc and adh. While bibp was successfully over-expressed, the amount of BiBP was much higher in strains containing the pDF-BiBP constructs. Further studies with ethanol producers should be done by transforming EE with the pDF-BiBP constructs to increase bibp expression levels.
The original idea behind this project was to explore the effect of both light intensity and [CO2] on growth and ethanol production in BiBP-overexpressing strains. However, due to both time constraints and the breakdown of two Algaetron incubators, cultivation was only able to be done under high light intensity at atmospheric [CO2]. Future experiments would include completing the original purpose of this study and run growth analysis under high [CO2] alongside high light intensity, and compare growth and ethanol production to strains cultured under low light intensity and atmospheric [CO2]. If the results of this current study are because of RuBP regeneration not being the major limiting step due to the atmospheric [CO2], repeating the experiments under high [CO2] should give positive results.
Cell size relative to wild type strain should also be measured for the strains in this work and any subsequent work. Euglena gracilis overexpressing BiBP were shown to have an increased cell volume but not growth rate under atmospheric [CO2]; both cell volume and growth rate were increased relative to the wild type under elevated [CO2] [23]. Increased cell volume in BiBP-overexpressed 7002 strains was also reported by De Porcellinis et al. Since it is possible for cultures to exhibit an increased cell size without an increase in optical density, it could be possible that the cells in this work have indeed experienced an increase in growth in the form of increased cell size but not doubling time. Dry weight of cultures could also be measured as another indicator of increased growth other than optical density.
7002 strains grown under 1000 μE light intensity at a starting OD of 0.2-0.4 showed signs of photobleaching after 3 days of cultivation. This was unexpected, as 7002 is known to be resistant to high light intensities, and photobleaching was not reported for strains cultivated under 1500 μE light intensity in the work of De Porcellinis et al. Photobleaching was also not reported by Dexter and Fu in their work where ethanol-producing 6803 strains were cultured in a BioFlo 110 bioreactor system under 1000 μE [1]. This inconsistency could be due to the culture volumes involved, since the strains in the work of Dexter and Fu were cultured at 3.1 L. However, since the MC1000 only allows a maximum volume of 80 mL per culture, strains should be grown up to a higher initial OD before cultivation under high light intensity in future work.
Several other phenotypes reported by other labs should also be investigated. The regulatory effect of BiBP on other enzymes of carbon metabolism, especially, Rubisco, should be explored using SRM proteomics. An interesting question would be whether there is a linear relationship between increase in BiBP expression and activation state, amount, and activity of Rubisco; this would go a long way towards clarifying the interaction between BiBP and Rubisco. De Porcellinis et al. also reported the repression of enzymes involved in carbon catabolism, results that have not been confirmed by other studies. It would be worth validating these claims by using SRM proteomics to quantify the amounts of these enzymes relative to the control.
References
Dexter, J. and Fu, P. (2009). Metabolic engineering of cyanobacteria for ethanol production. Energy & Environmental Science, 2(8), p.857.
Gao, Z., Zhao, H., Li, Z., Tan, X. and Lu, X. (2012). Photosynthetic production of ethanol from carbon dioxide in genetically engineered cyanobacteria. Energy Environ. Sci., 5(12), pp.9857-9865.
Dexter, J., Armshaw, P., Sheahan, C. and Pembroke, J. (2015). The state of autotrophic ethanol production in Cyanobacteria. Journal of Applied Microbiology, 119(1), pp.11-24.
Feng, L., Sun, Y., Deng, H., Li, D., Wan, J., Wang, X., Wang, W., Liao, X., Ren, Y. and Hu, X. (2013). Structural and biochemical characterization of fructose-1,6/sedoheptulose-1,7-bisphosphatase from the cyanobacteriumSynechocystisstrain 6803. FEBS Journal, 281(3), pp.916-926.
Tamoi, M., Murakami, A., Takeda, T. and Shigeoka, S. (1998). Acquisition of a new type of fructose-1,6-bisphosphatase with resistance to hydrogen peroxide in cyanobacteria: molecular characterization of the enzyme from Synechocystis PCC 6803. Biochimica et Biophysica Acta (BBA) – Protein Structure and Molecular Enzymology, 1383(2), pp.232-244.
Jiang, Y., Wang, D. and Wen, J. (2012). The independent prokaryotic origins of eukaryotic fructose-1, 6-bisphosphatase and sedoheptulose-1, 7-bisphosphatase and the implications of their origins for the evolution of eukaryotic CBB Cycle. BMC Evolutionary Biology, 12(1), p.208.
Zhu, X., de Sturler, E. and Long, S. (2007). Optimizing the Distribution of Resources between Enzymes of Carbon Metabolism Can Dramatically Increase Photosynthetic Rate: A Numerical Simulation Using an Evolutionary Algorithm. PLANT PHYSIOLOGY, 145(2), pp.513-526.
Raines, C. (2003). The Calvin Cycle Revisited. Photosynthesis Research, 75: 1–10.
Poolman, M., Fell, D. and Thomas, S. (2000). Modelling photosynthesis and its control. Journal of Experimental Botany, 51(suppl_1), pp.319-328.
Farquhar, G., von Caemmerer, S. and Berry, J. (1980). A biochemical model of photosynthetic CO2 assimilation in leaves of C3 species. Planta, 149(1), pp.78-90.
Raines, C. (2010). Increasing Photosynthetic Carbon Assimilation in C3 Plants to Improve Crop Yield: Current and Future Strategies. PLANT PHYSIOLOGY, 155(1), pp.36-42.
Raines, C. (2006). Transgenic approaches to manipulate the environmental responses of the C3 carbon fixation cycle. Plant, Cell and Environment, 29(3), pp.331-339.
Ding, F., Wang, M., Zhang, S. and Ai, X. (2016). Changes in SBPase activity influence photosynthetic capacity, growth, and tolerance to chilling stress in transgenic tomato plants. Scientific Reports, 6(1).
Driever, S., Simkin, A., Alotaibi, S., Fisk, S., Madgwick, P., Sparks, C., Jones, H., Lawson, T., Parry, M. and Raines, C. (2017). Increased SBPase activity improves photosynthesis and grain yield in wheat grown in greenhouse conditions. Philosophical Transactions of the Royal Society B: Biological Sciences, 372(1730), p.20160384.
Lefebvre, S. (2005). Increased Sedoheptulose-1,7-Bisphosphatase Activity in Transgenic Tobacco Plants Stimulates Photosynthesis and Growth from an Early Stage in Development. PLANT PHYSIOLOGY, 138(1), pp.451-460.
Raines, C., Harrison, E., Ölçer, H. and Lloyd, J. (2008). Investigating the role of the thiol-regulated enzyme sedoheptulose-1,7-bisphosphatase in the control of photosynthesis. Physiologia Plantarum, 110(3), pp.303-308.
Rosenthal, D., Locke, A., Khozaei, M., Raines, C., Long, S. and Ort, D. (2011). Over-expressing the C3 photosynthesis cycle enzyme Sedoheptulose-1-7 Bisphosphatase improves photosynthetic carbon gain and yield under fully open air CO2 fumigation (FACE). BMC Plant Biology, 11(1), p.123.
Simkin, A., Lopez-Calcagno, P., Davey, P., Headland, L., Lawson, T., Timm, S., Bauwe, H. and Raines, C. (2017). Simultaneous stimulation of sedoheptulose 1,7-bisphosphatase, fructose 1,6-bisphophate aldolase and the photorespiratory glycine decarboxylase-H protein increases CO2 assimilation, vegetative biomass and seed yield in Arabidopsis. Plant Biotechnology Journal, 15(7), pp.805-816.
Harrison, E., Olcer, H., Lloyd, J., Long, S. and Raines, C. (2001). Small decreases in SBPase cause a linear decline in the apparent RuBP regeneration rate, but do not affect Rubisco carboxylation capacity. Journal of Experimental Botany, 52(362), pp.1779-1784.
Miyagawa, Y., Tamoi, M. and Shigeoka, S. (2001). Overexpression of a cyanobacterial fructose-1,6-/sedoheptulose-1,7-bisphosphatase in tobacco enhances photosynthesis and growth. Nature Biotechnology, 19(10), pp.965-969.
Tamoi, M., Nagaoka, M., Miyagawa, Y. and Shigeoka, S. (2006). Contribution of Fructose-1,6-bisphosphatase and Sedoheptulose-1,7-bisphosphatase to the Photosynthetic Rate and Carbon Flow in the CBB Cycle in Transgenic Plants. Plant and Cell Physiology, 47(3), pp.380-390.
Köhler, I., Ruiz-Vera, U., VanLoocke, A., Thomey, M., Clemente, T., Long, S., Ort, D. and Bernacchi, C. (2016). Expression of cyanobacterial FBP/SBPase in soybean prevents yield depression under future climate conditions. Journal of Experimental Botany, p.erw435.
Ogawa, T., Tamoi, M., Kimura, A., Mine, A., Sakuyama, H., Yoshida, E., Maruta, T., Suzuki, K., Ishikawa, T. and Shigeoka, S. (2015). Enhancement of photosynthetic capacity in Euglena gracilis by expression of cyanobacterial fructose-1,6-/sedoheptulose-1,7-bisphosphatase leads to increases in biomass and wax ester production. Biotechnology for Biofuels, 8(1).
Ichikawa, Y., Tamoi, M., Sakuyama, H., Maruta, T., Ashida, H., Yokota, A. and Shigeoka, S. (2010). Generation of transplastomic lettuce with enhanced growth and high yield. GM Crops, 1(5), pp.322-326.
Feng, L., Han, Y., Liu, G., An, B., Yang, J., Yang, G., Li, Y. and Zhu, Y. (2007). Overexpression of sedoheptulose-1,7-bisphosphatase enhances photosynthesis and growth under salt stress in transgenic rice plants. Functional Plant Biology, 34(9), p.822.
Gong, H., Li, Y., Fang, G., Hu, D., Jin, W., Wang, Z. and Li, Y. (2015). Transgenic Rice Expressing Ictb and FBP/Sbpase Derived from Cyanobacteria Exhibits Enhanced Photosynthesis and Mesophyll Conductance to CO2. PLOS ONE, 10(10), p.e0140928.
Liang, F. and Lindblad, P. (2016). Effects of overexpressing photosynthetic carbon flux control enzymes in the cyanobacterium Synechocystis PCC 6803. Metabolic Engineering, 38, pp.56-64.
Lutz, R. (1997). Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Research, 25(6), pp.1203-1210.
Guerrero, F., Carbonell, V., Cossu, M., Correddu, D. and Jones, P. (2012). Ethylene Synthesis and Regulated Expression of Recombinant Protein in Synechocystis sp. PCC 6803. PLoS ONE, 7(11), p.e50470.
Camsund, D., Heidorn, T. and Lindblad, P. (2014). Design and analysis of LacI-repressed promoters and DNA-looping in a cyanobacterium. Journal of Biological Engineering, 8(1), p.4.
Markley, A., Begemann, M., Clarke, R., Gordon, G. and Pfleger, B. (2014). Synthetic Biology Toolbox for Controlling Gene Expression in the Cyanobacterium Synechococcus sp. strain PCC 7002. ACS Synthetic Biology, 4(5), pp.595-603.
Cite This Work
To export a reference to this article please select a referencing stye below:
Related Services
View allRelated Content
All TagsContent relating to: "Environmental Studies"
Environmental studies is a broad field of study that combines scientific principles, economics, humanities and social science in the study of human interactions with the environment with the aim of addressing complex environmental issues.
Related Articles
DMCA / Removal Request
If you are the original writer of this dissertation and no longer wish to have your work published on the UKDiss.com website then please:




