The Genus Macrococcus: Biology, Evolution and Relationship with Staphylococcus
Info: 8773 words (35 pages) Dissertation
Published: 9th Dec 2019
Tagged: Biology
The Genus Macrococcus: An insight into its biology, evolution and relationship with Staphylococcus.
Contents
1. Abstract
2. Introduction
3. Description of the Macrococcus Genus
3.1 Taxonomic History
3.2 Phylogeny
3.2.1 Identification-General properties of Macrococcus Genus
3.2.2 Methods-Differentiation of species
3.2.3 Description of the species
3.2.3.1 Macrococcus caseolyticus
3.2.3.2 Macrococcus equipercicus
3.2.3.3 Macrococcus bovicus
3.3.3.4 Macrococcus carouselicus
3.2.3.5 Macrococcus brunensis
3.2.3.6 Macrococcus hajekii
3.2.3.7 Macrococcus lamae
3.2.3.8 Macrococcus canis
4. Differentiation of Macrococcus species from Staphylococcus
4.1 Cell wall composition
4.2 Phenotypic Characteristics
4.3 DNA-DNA hybridisation and DNA base composition
4.4 Other Molecular Methods
5. Antibiotic resistance
6.1 Dissemination of Methicillin resistance
6.2 Mechanisms of Multidrug resistance species of Macrococcus.
7. Application
8. References
Advances in Applied Microbiology,
Abstract
Introduction
Macrococcus species disseminated in nature as animal commensals, after the divergence of its ancestral bacterium from the family Bacillaceae (Baba et al., 2009). Macrococcal species are direct antecedents of staphylococcal species (Hiramatsu et al., 2014) and the genus is classified taxonomically in the family of Staphylococcaceae (Lory, 2014). Currently eight species exist in the Macrococcus genus: Macrococcus caseolyticus, Macrococcus bovicus, Macrococcus carouselicus, Macrococcus equipercicus, Macrococcus brunensis, Macrococcus hajekii, Macrococcus lamae and Macrococcus canis (Brawand, et al., 2017) The species belonging to Macrococcus genus are Gram positive, oxidase and catalase positive cocci, that occur in pairs, tetrads or in irregular clusters (Schleifer, 2015)
Description of the Macrococcus Genus
Taxonomic History: Staphylococcus and Macrococcus
Historically many classifications of the Gram-positive and catalase-positive cocci have been suggested, in 1820’s researcher first observed cocci shaped bacteria present in inflammation, abscesses and pus (Hill, 1959). Toxicogenic properties of these cocci were described in a classical paper by Ogston “ Micrococcus Poisoning” (Kitching and Farrell, 1936), he was first to introduce the name Staphylococcus by differentiating spherical bacteria in to two types (Ogston, 1882), those arranged in groups of clusters were called Staphylococcus and others in chains were referred to as Streptococcus (Ogston, 1882). In 1884 Rosenbach provided a formal taxonomical classification of the Staphylococcus genus, when he divided the genus in to two species Staphylococcus aureus and Staphylococcus albus (Rosenbach, 1884). These staphylococci and tetrad forming micrococci were deposited in to the genus Micrococcus by Zopf (Zopf, 1885). The first species of Macrococcus was primarily isolated from raw milk in 1916 by Evan and was placed in to the Micrococcus genus based on the morphological and carbohydrates decomposition characteristic’s (Evans, 1916). Developments in the area of simple fermentation and oxidation test resulted in distinguishing Micrococcus from Staphylococcus (Baird-Parker, 1965). These traditional approaches were applied by Flugge, Evans, Bradford & Niven results indicated staphylococci to produce gelatinises and acid anaerobically from glucose (Flügge, 1886, Evans et al., 1955) (Baird-Parker, 1963). However these test were not sufficient to differentiate weak anaerobic staphylococci from certain micrococci (SCHLEIFER et al., 1982b) but with the major advancement in DNA base composition a clear distinction between these two Gram positive cocci could be made. This demonstrated that members of the Staphylococcus genus have a lower per cent G+C content of 33–40 mol %, whereas members of the Micrococcus genus had a higher per cent G+C content of DNA around 63-73 mol % (Silvestri and Hill, 1965). Further studies have illustrated differentiation and identification of staphylococci from micrococci and other bacteria on the basis of their cell wall composition (Schleifer and Kandler, 1972) cytochrome and menaquinone patterns (Faller et al., 1980, Collins and Jones, 1981), sensitivity to erythromycin, lysostaphin, bacitracin and furazolidone (Schleifer and Kloos, 1975, Falk and Guering, 1983, Baker, 1984). Polar lipid composition (Nahaie et al., 1984) DNA-rRNA hybridization (Kilpper et al., 1980), and comparative oligonucleotide cataloguing of 16S rRNA (LUDWIG et al., 1981). Later comparative, chemical biochemical and nucleic acid hybridisation analyses were performed on three Micrococcus strains including Micrococcus caseolyticus ATCC13548, which was then assigned to the genus Staphylococcus on the basis of its cell wall composition , and low G+C content of DNA and DNA-rRNA hybridization. This strain was distinct from the other known members of the Staphylococcus, and proposed to represent new species of this genus and referred to as Staphylococcus caseolyticus (SCHLEIFER et al., 1982b). Staphylococcus caseolyticus was then reclassified to a newly proposed genus Macrococcus in 1998 by Kloos, this genus was described independent and separate from its closest relative Staphylococcus on the basis of its comparative 16S rRNA analysis, DNA-DNA hybridization, ribotype patterns, cell wall composition, and phenotypic characteristics (Kloos et al., 1998a). Member of the Macrococcus genus can be distinguished from Staphylococcus on the basis of its larger cell size, higher G+C content 38–45 mol %, smaller genome size and significantly lower 16S rRNA sequence similarity. Macrococci are also oxidase positive whereas majority staphylococci are negative (exceptions: Staphylococcus sciuri, S. lentus, S. pulvereri and S. vitulus). Currently eight species exist in the Macrococcus genus: Macrococcus caseolyticus, Macrococcus bovicus, Macrococcus carouselicus, Macrococcus equipercicus, Macrococcus brunensis, Macrococcus hajekii, Macrococcus lamae and Macrococcus canis (Brawand et al., 2017). Macrococcus genus now belongs in the family of Staphylococcaceae (Lory, 2014) and its 16 S rRNA analysis indicates the closest relative to be Staphylococcus and also clusters with other Gram positive bacteria with low DNA G+C content such as Salinicoccus and Bacillus (Kloos et al., 1998a, Götz et al., 2006).
Phylogeny
On the basis of comparative 16S rRNA gene sequence studies, the genus Macrococcus belongs to the phylum of Firmicutes which consists of low DNA G+C content Gram positive bacteria. They are most closely related to Staphylococcus and other members of the Firmicutes such as members of the family of Listeriaceae and other Gram positive bacteria with low DNA G+C content such as bacilli, enterococci, streptococci and lactobacilli (Fig. 1) (Vos et al., 2011). Taxonomically the genus is place in to the family of Staphylococcaceae which was proposed to combine the genera Staphylococcus, Gemella, Macrococcus and Salinicoccus (Lory, 2014). Currently the Staphylococcaceae family also includes the species from Jeptgalicoccus and Nosocomiicoccus (ref hsp)60.
The genera Staphylococcus and Macrococcus are monophyletic (Fig. 2) and seperated from each other with intergenera 16S rRNA sequence similarities of 93.4–95.3% whereas intragenus similarities of staphylococci is at least 96.5% and slightly higher for micrococci which is 97.7% (Vos et al., 2011). Phylogenetic study of Staphylococcus species and four of the eight Macrococcus species (M. equipercicus, M. bovicus, M. carouselicus and M. caseolyticus) based on comparative sequence analysis of heat-shock protein (hsp60) indicates clear separation from the Staphylococcus species but cluster closely to the group of Staphylococcus species belonging to the S. sciuri group the only species of Staphylococcus that produce cytochrome c oxidase (S. sciuri, S. lentus, S. pulvereri and S. vitulus) which is also evident in the 16S rRNA sequence analysis from a number of studies( family and delimination).The pairwise sequence identity scores based on partial amplification of hsp60 gene were significantly lower than 16S rRNA, for four macrococcal species ranged from 82-87%, while those amongst the 40 staphylococcal species ranged from74-98%. Thereby, hsp60 gene sequence appears to be more discriminatory than the 16S rRNA gene sequence for identification and differentiation of staphylococcal and macrococcal species (Kwok and Chow, 2003).
DNA-DNA based studies of the four Macrococcus species indicated M. equipercicus, M. bovicus, M. carouselicus were more closely related to one another than M. caseolyticus (Kloos et al., 1998a). All of the studies based on DNA-DNA hybridization, partial hsp60 gene and 16S rRNA sequence analysis indicate a closer relationship of the genus Macrococcus to the S. sciuri species group than to other staphylococcal species outside this group. Particularly, M. caseolyticus and M. canis are more closely related to Staphylococcus species than the other Macrococcus species, this correlates with significantly lower G+ C content (36.9-38 mol%) of the M. caseolyticus and M. canis that is shared by several staphylococcal species.
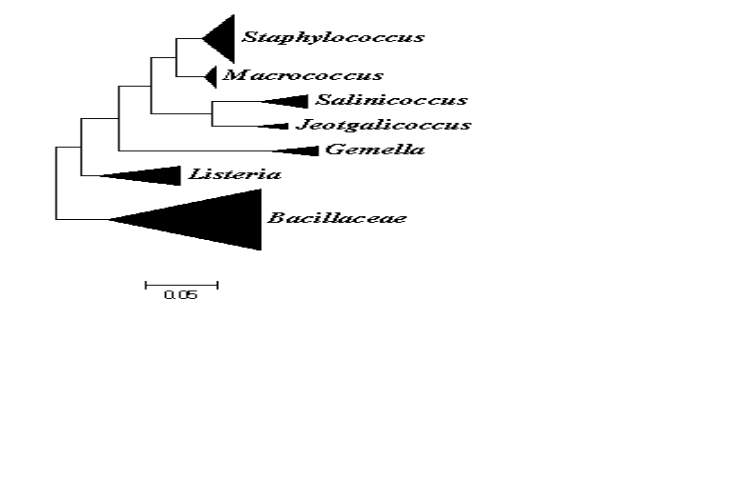
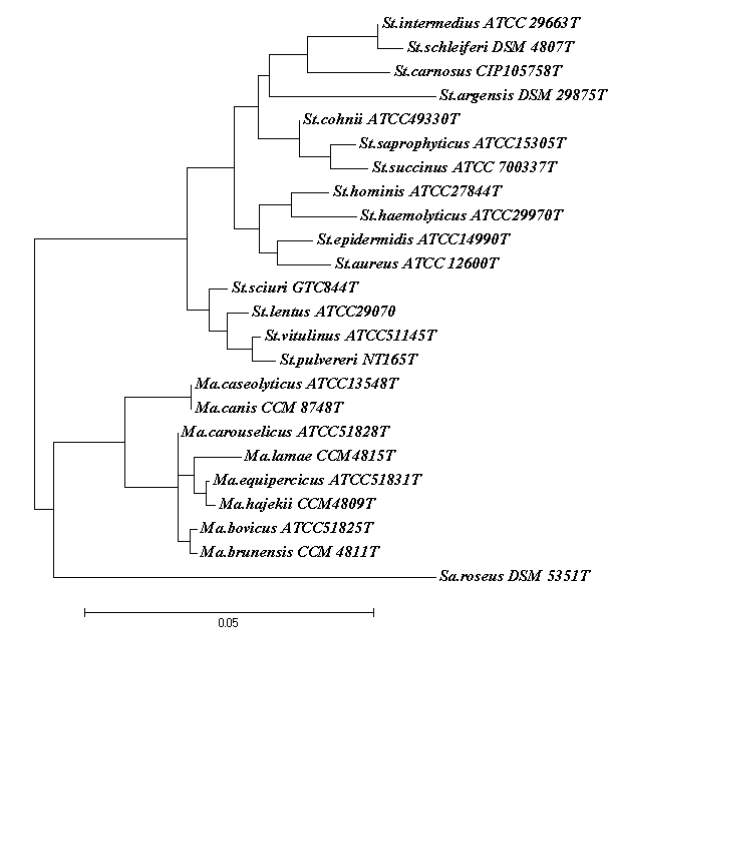
Identification
General Properties of Macrococcus Genus
The name Macrococcus “large coccus” (Kloos et al., 1998a) was adopted, as the members of this genus have larger cell size in contrast to the species of its sister genus Staphylococcus (cell size ≈ 2.5-4 times the diameter of Staphylococcus aureus cells when grown in TS broth). The macrococcalspecies are Gram positive, that occur mostly in pairs, tetraheards or in irregular clusters and occasionally single or arranged in short chains (Kloos et al., 1998a, Götz et al., 2006). Cells are spherical or coccoid shaped (1.1-2.5 µm in diameter). They are generally unencapsulated, non-motile and non-sporeforming. Macrococci are chemoorganotrophic, metabolisim is predominantly respiratory; marginally facultatively anaerobic; grow better under aerobic conditions. They are coagulase negative and catalase and cytochrome c oxidase positive (Götz et al., 2006). They are negative for ornithine decarboxylase, β-glucuronidase, arginine deiminase activities and for acid production from the fermentation of D-cellobiose, D-melezitose, D-raffinose , D-turanose, D-xylose, L-arabinose and salicin. Macrococcalspecies are generally more susceptible to a wide range of antibiotics in comparison to staphylococcal species which includes: penicillin G, erythromycin, clindamycin, tetracycline, ciprofloxacin, rifampin, trimethoprim-sulfamethoxazole, gentamicin, kanamycin, streptomycin, vancomycin and chloramphenicol (Kloos et al., 1998a). However, macrococci are resistant to bacitracin and lysozyme, and susceptible to furazolidone. They contain a-, b- and/or c type cytochromes whereas majority of the staphylococci contain a- and b- type cytochromes with the exception of S. sciuri group (S. sciuri, S. lentus, S. pulvereri and S. vitulus) that possess c type cytochromes. The cell wall composition of macrococci with the exception of M. caseolyticus do not contain teichoic acid, lipoteichoic acid is present in M. caseolyticus, M. carouselicus, M. equipercicus and M. boviscus. Peptidoglycan types are either Lys-Gly3-4 , L-Ser (M. caseolyticus, M. carouselicus and M. equipercicus) or Lys-Gly3, L-Ser (M. bovicus) (Kloos et al., 1998a). The genome size range is approximately 1500-1800 kb. The G + C content of the DNA is 36.9-45 mol% (Mannerová et al., 2003, Brawand et al., 2017).
Cell Wall composition
As there is diversity in the structure of the peptidoglycan layer in Gram positive bacteria, for this reason the cell wall structure analysis and chemical composition plays an important taxonomic criterion in the differentiation of these bacterium. Methods utilised to isolate peptidoglycans and elucidate its structure dates back to the 1960’s (Ghuysen et al., 1968, Schleifer and Kandler, 1972). These methods along with other methods were used to analyse lipoteichoic (Ruhland and Fiedler, 1990) and teichoic acids(Anderson et al., 1977) composition in the cell wall of strains belonging to M. caseolyticus M. equipercicus, M. bovicus and M.carouselicus species (Kloos et al., 1998a). The peptidoglycan for these species is determined to be of A3α (L-Lys-Gly3-4,- L-Ser) type , which is a common type that is also found amongst several staphylococci species(Kloos et al., 1998b). Recently proposed species M. canis cell wall peptidoglycan analysis of its typed strain KM 45013T also indicate A3α type peptidoglycan (L-Lys–Gly2–L-Ser )(Brawand et al., 2017). However variations are identified in the levels of L-serine in the cell wall of certain Macrococcus species, highest level are reported to be present in the strains of M. caseolyticus (1-2-1.3 mol-1) where lowest levels are identified in the strains of M. bovicus (0.44-0.58 mol-1). The cell wall teichoic acids are documented to present solely in M. caseolyticus species. These copolymers are generally found to be present in all of the staphylococcal species(Endl et al., 1983), however the teichoic acid present in M. caseolyticus is in lower amount and is atypical of the type poly(N-acetylglucosaminylphosphate). Presence of lipoteichoic acid is identified to be present at the cell surface of M. caseolyticus, M. equipercicus, M. bovicus and M. carouselicus species. The glycolipid in the cell membrane contains glycosyl residues of gentiobiosyl, which are same as glycosyl that’s is found in staphylococci (Kloos et al., 1998a).
Methods-Differentiation of Macrococcus species
The classification and identification of species of macrococci is based on a variety of comparative phenotypic, chemotaxonomic and genotypic analysis. DNA-DNA hybridisation, DNA-rRNA hybridization ribotyping and 16S rRNA sequencing methods have been used extensively to elucidate phylogenetic position and differentiate these species from the species of staphylococci (Schleifer et al., 1982a, Kloos et al., 1998a, Mannerová et al., 2003). Established conventional methods were exploited in the 1980’s by Schleifer et al (Schleifer et al., 1982a) to study characters at the cellular and population, included morphological and physiological properties (Baird-Parker, 1963), cell wall composition (Schleifer and Kandler, 1972, Schleifer, 1973), menaquinone pattern (Jeffries et al., 1968), cytochrome pattern (Faller et al., 1980), enzyme reactions (Götz et al., 1979, Faller and Schleifer, 1981), sensitivity to lysostaphin (Schleifer and Kloos, 1975). These phenotypic characteristics were combined with additional characteristics that were investigated by other studies (Kloos et al., 1998a) and examined for their correlation with DNA-DNA (genomic) hybridisation (SCHLEIFER et al., 1982b), DNA base composition and 16S rRNA sequencing, fatty acid and methyl esters (FAME) analysis (Mannerová et al., 2003), ribotype and macrorestriction patterns to identify macrococci independent from staphylococci as well as distinguishing species of Macrococcus from one another.
Key characters now used to identify and differentiate species of Macrococcus include the following: cell morphology and arrangement, colony morphology and pigmentation, acetoin production, nitrate reduction, oxidase activity, aesculin hydrolysis, DNase activity, thermonuclease activity, urease activity, phosphatase production, lysostaphin, novobiocin and oxacillin susceptibility, haemolysis, CAMP reaction and aerobic acid production from variety of carbohydrates including D-mannitol, D-mannose, D-sorbitol, D-ribose, maltose, sucrose, lactose and glycerol (Kloos et al., 1998a, Mannerová et al., 2003, Brawand et al., 2017).
Description of the species from the genus of Macrococcus
Macrococcus caseolyticus
First description of this species was made in the 1916 by Evans, who isolated this bacterium from cow’s milk. This species was characterised by its ability to rapidly and completely peptonise the milk and therefore, the name caseolyticus was suggested which means “casein-dissolving”(Evans, 1916). M. caseolyticus previously referred to as Staphylococcus caseolyticus was investigated by ballard for its presence in artiodactyl and cetacean host. Investigation identified M. caseolyticus species as relatively uncommon as strains of this species were isolated at lower population from the skin of cattle, sheep, goats and pilot skin whale (Kloos et al., 1998a), in contrast to other species of Macrococcus which were identified in larger amounts to be present in similar sources (except cetacean host). Other studies have reported presence of this species in milk (Evans, 1916, Schwendener et al., 2017) and meat products (Baba et al., 2009, Wu et al., 2009)(Kloos et al., 1998a). General characteristics of this species have been delineated in a number of studies. Few of which include the following: The cells are spherical, 1.1-2µm in diameter. Colonies are pigmented; pale yellow or unpigmeneted; white-grey, slightly halotolerant (10%NACL)(Mannerová et al., 2003), negative for β-haemolysis (sheep, horse and bovine blood)(Kloos et al., 1998a) and coagulase production, produces small amount of L-(+)-lactic acid from fermentation of glucose (SCHLEIFER et al., 1982b), cell wall contains atypical teichoic acid (Götz et al., 2006), produceS class II fructose- 1,6-diphosphate aldolase (SCHLEIFER et al., 1982b),reduces nitrates, negative urease and β-glucosidase activities, no acid production from D-mannitol, D-cellobiose, L-arrabinose and glycerol, produces acetoin and strains of M. caseolyticus are capable of hydrolysing casein. The G+C content of the DNA of the two fully sequenced strains JCSC5402 and IMD1089 of M. caseolyticus is 36.5-36.9% whereas previous studies have reported G+C content of the DNA to be slightly higher 38-39%(KLOOS AND SCH).
Macrococcus equipercicus
sp. nov. Macrococcus equipercicus (equi.per’ci.cus. L. gen. n. equi of a horse. M.L. adj. equipercicus pertaining to a horse named Percy, from which this species was first isolated).
The description below is based on the characteristics of 22 strains (Table 1) isolated in 1993-1995 from the skin of horses and ponies. Colonies grow to 6+2 mm in diameter on P agar and 6+ 1 mm on TSA. They are convex, entire, butyrous, dull to slightly glistening and opaque, and have a light- to medium-orange pigmentation. Growth is not detected in the anaerobic portion of a semi-solid thioglycollate medium. Growth is good at NaCl concentrations up to 7.5 YO. Optimum growth temperature is 35 “C. Culture growth does not cause haemolysis of horse, bovine or sheep blood. The cell wall does not contain teichoic acid. Acetoin is not produced and nitrates are not reduced. DNase, pyrrolidonylarylamidase and #?-glucosidase activities are negative, except that strain DD 9350T is weakly positive for DNase and /3-glucosidase activities. Staph Latex agglutination is negative, except that strain DD 11654 is weakly positive for this test. All strains are resistant to novobiocin and susceptible to lysostaphin. Acid is produced aerobically from glycerol and #?-Dfructose. All strains, except DD 9347, produce acid from D-mannitol. Acid is not produced from a-lactose and, with the exception of strain DD 9347, is not produced from D-ribose. Characteristics of M. equipercicus that are general properties of the genus are listed above. The variable characteristics for this species are listed in Table 4. The major API STAPHIDENT profile is 2000 (68 YO). There are ten different ID32 STAPH profiles; the most common ones begin with the digits 17 (27 YO) or 35 (1 8 YO) in the nine-digit profile, although no specific profile is represented by more than 14% of the strains. The major ribotype pattern types are dd 11639 (41 YO) and dd 09345 (23 YO). The G + C content of the DNA is 45 mol YO. This species has a preference for perissodactyls (e.g. horses and ponies) and is commonly found as large populations on the skin of these mammals.
Macrococcus bovicus
sp. nov. Macrococcus bovicus (bov.ic’us. Gr. n. bou cow. L. gen. n. bovis of a cow. M.L. adj. bovicus pertaining to a bovine or cow, from which this organism was first isolated). The description below is based on the characteristics of ten strains (Table 1) isolated in 1992-1994 from the skin of a cow, a pony and several horses. Colonies grow to 4+ 1 mm in diameter on P agar and TSA. They are slightly convex, entire, butyrous, glistening and opaque, and have a pale-yellow to medium-orange pigmentation. Growth is not detected in the anaerobic portion of a semi-solid thioglycollate medium. Growth is good at NaCl concentrations up to 7.5 YO. Optimum growth temperature is 35 “C. Culture growth causes a partial haemolysis (greening) of horse and bovine blood. The cell wall does not contain teichoic acid. Lipoteichoic acid is present. Acetoin is not produced. Pyrrolidonylarylamidase activity is negative. All strains are resistant to novobiocin and susceptible to lysostaphin and oxacillin. Acid is produced aerobically from D-mannitol, glycerol and P-D-fructose. Acid is not produced from a-lactose. Characteristics of M. bovicus that are general properties of the genus are listed above. The variable characteristics of this species are listed in Table 4. Major API STAPH-IDENT profiles are 0200 (30 %), 4500 (20 YO) and 4600 (20 %), and major ID32 STAPH profiles are 062200000 (20 %), 0623 10200 (20 %) and 072300200 (20 YO). The major ribotype pattern type is dd 09344 (40%). The G+C content of the DNA is 42-44 mol%. This species appears to have a preference for perissodactyls (e.g. horses and ponies) and artiodactyls (e.g. cattle). Description of the type strain. Type strain is ATCC 51825T (= DD 4516T), isolated from the skin of a Holstein cow. It has all of the general properties of the species and genus described above and also the following characteristics. Cells are spherical, 1.2- 2.1 pm in diameter, have a slightly irregular surface (SEM), and occur singly and in pairs, tetrads and short chains. Colonies on P agar and TSA are circular, 3-5 mm in diameter, entire, slightly convex, and with a glistening surface and butyrous consistency. Mediumorange pigmentation. Marginally facultatively anaerobic; no visible growth in anaerobic portion of thioglycollate medium. Partial haemolysis (greening) of horse and bovine blood. Positive DNase activity and latex agglutination. Negative aesculin hydrolysis, nitrate reduction and urease activity. Acid produced aerobically from D-trehalose, D-mannitol, glycerol and p-D-fructose. No acid produced from sucrose, alactose, maltose and D-ribose. Type dd 04516 ribotype pattern. Cell wall peptidoglycan is ~-Lys-Gly,, L-Ser. No detectable teichoic acid. Lipoteichoic acid is present. G + C content of DNA is 44 mol %.
Macrococcus carouselicus
(car.ou.se1’i.cus. M.L. adj. carouselicus pertaining to a carousel or merry-goround, which has carousel horses). The description below is based on the characteristics of 18 strains (Table 1) isolated in 1993-1995 from the skin of horses and ponies. Colonies grow to 5 + 2 mm in diameter on P agar and 7& 1 mm on TSA. They are slightly convex, entire, butyrous, glistening and opaque, and have a cream- to light-orange pigmentation. Growth is not detected in the anaerobic portion of a semi-solid thioglycollate medium. Growth is good at NaCl concentrations up to 7.5 %. Optimum growth temperature is 35 “C. Culture growth usually does not cause haemolysis of horse and sheep blood. The cell wall does not contain teichoic acid. Lipoteichoic acid is present. Acetoin is not produced and, with the exception of strain DD 9346, nitrates are not reduced. All strains, except DD 934 1, hydrolyse aesculin. DNase activity is positive. All strains, except DD 9607, are negative for P-glucosidase activity. Pyrrolidonylarylamidase activity is negative. All strains are resistant to novobiocin and susceptible to lysostaphin and oxacillin. All strains, except DD 9341, weakly produce acid aerobically from P-D-fructose. Acid is not produced from maltose, D-ribose and a-lactose. Characteristics of M. carouselicus that are general properties of the genus are listed above. The variable characteristics of this species are listed in Table 4. The major API STAPH-IDENT profile is 0000 (94 YO) and major ID32 STAPH profiles are 050100000 (28%), 070100000 (28%) and 010100000 (22%). The major ribotype pattern types are dd 09342 (28%) and dd 09349 (17%). The G+C content of the DNA is 41 mol %. This species has a preference for perissodactyls (e.g. horses and ponies) and is commonly found as large populations on the skin of these mammals.
Macrococcus brunensis
sp. nov. Macrococcus brunensis (bru.nen9sis. L. adj. brunensis from Bruna, the Roman name of the city of Brno, Czech Republic, where the type strain was isolated). Colonies reach 2–4 mm in diameter on P agar after 24 h. Cell diameter is 0?89–1?21 mm. Colonies are circular, smooth and glossy, without pigment. Growth is detected under anaerobic conditions at 15–36 uC and in 4 % NaCl but not at 4 or 42 uC. All strains hydrolyse casein and gelatin, but not Tween 80, starch, lecithin, aesculin or tyrosine. Alkaline and acid phosphatases are produced, nitrates are reduced and acid is produced from maltose, D-mannitol, D-fructose, D-trehalose and D-glucose. Acetoin, clumping factor and coagulase are not produced and activities of urease, haemolysis, arginine dihydrolase, arginine arylamidase, ornithine decarboxylase, b-galactosidase, b-glucuronidase, pyrrolidonyl arylamidase, esterase (C4), lipase (C14), naphthol-AS-BI-phosphohydrolase, valine arylamidase, cystine arylamidase, a-galactosidase,N-acetyl-b-glucosaminidase, a-mannosidase, a-fucosidase and esterase-lipase (C8) are negative. Acid is not produced from sucrose, melezitose, xylose, arabinose, turanose, xylitol, D-sorbitol, D-cellobiose, D-salicin, raffinose, D-galactose, lactose, ribose, D-melibiose, D-mannose, N-acetylglucosamine or methyl a-D-glucoside. All strains are resistant to novobiocin. Predominant fatty acids are iso-13 : 0, iso-15 : 0, anteiso-15 : 0, 16 : 1v11c, iso-17 : 1v10c and 18 : 1v9c (Table 2). The G+C content of the DNA is 41–42 mol%. The type strain, CCM 4811T (=LMG 21712T ), was isolated from llama skin. Its characteristics are in full agreement with the species description. The G+C content of the DNA is 42 mol%.
Macrococcus hajekii
sp. nov. Macrococcus hajekii (ha.je9ki.i. N.L. gen. n. hajekii of Ha´jek, named after Va´clav Ha´jek, a Czech microbial taxonomist). Colonies reach 2–3 mm in diameter on P agar after 24 h. Mean cell diameter is 0?89 mm. Colonies are circular, smooth and glossy, without pigment. Does not grow at 4 or 42 uC or in 4 % NaCl, but grows at 15–36 uC. Hydrolyses casein and gelatin, but not Tween 80, starch, lecithin, aesculin or tyrosine. Alkaline phosphatase is produced, nitrates are reduced and acid is produced from sucrose, maltose, D-mannitol, D-fructose, D-trehalose and D-glucose. Acetoin, clumping factor and coagulase are not produced and activities of urease, haemolysis, arginine dihydrolase, argininearylamidase, ornithine decarboxylase,b-galactosidase, b-glucuronidase, pyrrolidonyl arylamidase, esterase (C4), lipase (C14), naphthol-AS-BI-phosphohydrolase, valine arylamidase, cystine arylamidase, a-galactosidase, Nacetyl-b-glucosaminidase, a-mannosidase, a-fucosidase, esterase-lipase (C8) and acid phosphatase are negative. Acid is not produced from melezitose, xylose, arabinose, turanose, D-sorbitol, D-cellobiose, xylitol, D-salicin, raffi- nose, D-galactose, lactose, ribose, D-melibiose, D-mannose, N-acetylglucosamine or methyl a-D-glucoside. Resistant to novobiocin. Dominant fatty acids are iso-13 : 0, iso-15 : 0, anteiso-15 : 0, 16 : 1v11c, iso-17 : 1v10c, 18 : 1v9c and 18 : 3v6c (Table 2). The G+C content of the DNA is 40 mol%. The type strain, CCM 4809T (=LMG 21711T ), was isolated from llama skin.
Macrococcus lamae
sp. nov. Macrococcus lamae(la9mae. N.L. fem. gen. n. lamae of Lama, the zoological genus name of the llama). Colonies reach 2–5 mm in diameter on P agar after 24 h. Cell diameter is 0?74–0?92 mm. Colonies are circular, smooth and glossy and have an orange pigment. All the strains tested grow at 15–36 uC. Growth is not detected under anaerobic conditions, in 4 % NaCl or at 4 or 42 uC. Strains hydrolyse casein and gelatin, but not Tween 80, starch, lecithin, aesculin or tyrosine. Alkaline phosphatase and esterase-lipase (C8) are produced. Acid is produced from sucrose, maltose, D-mannitol, D-fructose, D-trehalose and D-glucose. Acetoin, clumping factor and coagulase are not produced and activities of urease, haemolysis, arginine dihydrolase, arginine arylamidase, ornithine decarboxylase, b-galactosidase, b-glucuronidase, pyrrolidonyl arylamidase, esterase (C4), lipase (C14), naphthol-AS-BIphosphohydrolase, valine arylamidase, cystine arylamidase, a-galactosidase, N-acetyl-b-glucosaminidase, a-mannosidase, a-fucosidase are not observed. Nitrate is not reduced. Acid is not produced from melezitose, xylose, arabinose, turanose, Dsorbitol, D-cellobiose, D-salicin, raffinose, xylitol, D-galactose, lactose, ribose, D-melibiose, D-mannose,N-acetylglucosamine or methyl a-D-glucoside. Strain CCM 4812 does not produce acid phosphatase. Susceptible to novobiocin (5 mg) and intermediate susceptibility to novobiocin (1?6 mg). Predominant fatty acids are iso-13 : 0, 14 : 0, iso-15 : 0, anteiso15 : 0, ECL 15?669, 16 : 0, 16 : 1v11c, 18 : 0 and 20 : 0 (Table 2). The G+C content of the DNA is 41–42 mol%. The type strain, CCM 4815T (=LMG 21713T ), was isolated from llama skin. Its characteristics are in full agreement with the species description. The G+C content of the DNA is 41 mol%
Macrococcus canis
Application
It has been known that Gram positive, catalase positive and coagulase negative cocci play an important role in the development of characteristic flavour of fermented meat products (Martín et al., 2006, Kaban and Kaya, 2008). Flavour development of fermented foods involves complex network of metabolic reactions of which proteolysis plays a major role. In early literature M. caseolyticus referred to as Micrococcus caseolyticus has been documented to be present as part of secondary flora of cheese and its role in ripening due to its protease and peptidase activity has been documented (Bhowmik and Marth, 1988, Bhowmik and Marth, 1990). Moreno and Kosikowski have also investigated the activity of enzymes on β-casein from M. caseolyticus and other micrococci that lead to the production of short peptides and amino acids thereby suggesting that M. caseolyticus produce flavour compounds during cheese ripening (Moreno and Kosikowski, 1973). In dairy fermentation the proteolytic cascade begins with casein degradation by extracellular proteinases, M. caseolyticus ability to produce such extracellular caseinolytic enzyme has been described by Desmazeaud and Hermier (Desmazeaud and Hermier, 1968) and its activity on whole casein and β-casein has been delineated. Isolation of this extracellular enzyme described by Desmazeaud and Hermier (Desmazeaud and Hermier, 1968) from M. caseolyticus was exploited by a French company Roussel-Uclaf, who incorporated this protease in to liposomes for the production of a commercial enzyme called Rulactine (Yoovidhya et al., 1986). This metalloproteinase from M. caseolyticus was then used in the production of Saint Pauli cheese, its activity intensified proteolysis of milk and altered the texture of the curd (Alkalaf et al., 1987). Further investigations of adding Rulactine to the starter culture indicated significant acceleration of the cheese ripening process (Piard et al., 1986, Alkalaf et al., 1987, Alkhalaf et al., 2006). Technological application of enzymes extracted from a culture of this organism has also been described in a number of patents, the method for producing a polymeric enzyme from a culture of M. caseolyticus that is used in the production of aspartame which is a dipeptide that possess sweetening properties is described by the inventor Paul and Duchiron (Paul et al., 1990). Further exploitation of enzymes from a culture of M. caseolyticus and addition to milk had increased cheese capacity of milk used for the production of uncooked or half-cooked pressed paste cheeses (Barthelemy and Desmazeaud, 1986). M. caseolyticus was also used to produce novel cheese products and patents describe the technical contribution of the organism in the overall development of the desirable body and flavour of elastic cheese and low fat ripened skim milk cheese products (Hargrove and Mcdonough, 1964, Kasik and Luksas, 1971). M. caseolyticus conventional role in the flavour development of Cantonese sausage has also been investigated (Wu et al., 2009). Conventional role of M. caseolyticus in flavour and texture development in dairy and meat by producing functional biomolecules such as amino acids and volatile compounds have been investigated in the above studies. The ability of this organism to produce bioactive peptides was also investigated in one study where M. caseolyticus was screened for its inhibitory potential against shigatoxin-producing Escherichia coli (STEC) on a model cheese curd and results indicated this species to be amongst the most inhibitory (Callon et al., 2016) indicating it’s potential role in future bio-preservation. Another species from this genus; Macrococcus bovicus was also reported to be investigated for the development of an antibacterial technology (Abdel-Aziz et al., 2015). In this study M. bovicus used in the biosynthesis of nano-scaling silver (NSAg), this nano-composite illustrated to have an antimicrobial activity towards the Gram-negative and Gram-positive bacteria as well as fungus.
Table 1: List of species in the genera of Macrococcus.
Species name Source Referencea
Macrococcus caseolyticus Whale skin, Cow’s milk, Bovine tongue, Bovine heart, Food-processing factory (Kloos et al., 1998a)
Cow’s Milk (SCHLEIFER et al., 1982b)
M. boviscus Skin of Holstein cow, Irish thoroughbred horse, Morgan horse, Anglo-Trakehner horse (Kloos et al., 1998a)
M. equipercicus Skin of Irish thoroughbred horse, Morgan horse and Shetland pony (Kloos et al., 1998a)
M. carouselicus Skin ofIrish thoroughbred horse, Morgan horse, Anglo-Trakehner horse, Shetland pony (Kloos et al., 1998a)
M. brunensis Skin of llama’s (Mannerová et al., 2003)
M. hajekii Skin of llama’s (Mannerová et al., 2003)
M. lamae Skin of llama’s (Mannerová et al., 2003)
M. canis Canine Infection (Brawand et al., 2017)
a First description
References
Abdel-Aziz, M. S., K. S. Abou-El-Sherbini, E. M. A. Hamzawy, M. H. A. Amr, and S. El-Dafrawy. 2015. Green Synthesis of Silver Nano-particles by Macrococcus bovicus and Its Immobilization onto Montmorillonite Clay for Antimicrobial Functionality. Applied Biochemistry and Biotechnology 176(8):2225-2241.
Alkalaf, W., L. Vassal, M. Desmazeaud, and J. Gripon. 1987. Use of Rulactine as ripening agent in semi-hard cheese. Lait (France).
Alkhalaf, W., J.-C. Piard, M. El Soda, J. C. Gripon, M. Desmazeaud, and L. Vassal. 2006. Liposomes as Proteinase Carriers for the Accelerated Ripening of Saint‐Paulin Type Cheese. Vol. 53.
Anderson, A. J., R. S. Green, and A. R. Archibald. 1977. Specific determination of ribitol teichoic acid in whole bacteria and isolated walls of Bacillus subtilis W23. Carbohydrate research 57:C7-10.
Baba, T., K. Kuwahara-Arai, I. Uchiyama, F. Takeuchi, T. Ito, and K. Hiramatsu. 2009. Complete genome sequence of Macrococcus caseolyticus strain JSCS5402, reflecting the ancestral genome of the human-pathogenic staphylococci. Journal of bacteriology 191(4):1180-1190.
Baird-Parker, A. 1963. A classification of micrococci and staphylococci based on physiological and biochemical tests. Microbiology 30(3):409-427.
Baker, J. S. 1984. Comparison of various methods for differentiation of staphylococci and micrococci. Journal of clinical microbiology 19(6):875-879.
Barthelemy, P. and M. Desmazeaud. 1986. Process for preparing cheese and product produced. Google Patents.
Bhowmik, T. and E. H. Marth. 1988. Protease and peptidase activity of Micrococcus species. Journal of dairy science 71(9):2358-2365.
Bhowmik, T. and E. H. Marth. 1990. Rote of Micrococcus and Pediococcus Species in Cheese Ripening: A Review1. Journal of Dairy Science 73(4):859-866.
Brawand, S. G., K. Cotting, E. Gómez-Sanz, A. Collaud, A. Thomann, I. Brodard, S. Rodriguez-Campos, C. Strauss, and V. Perreten. 2017. Macrococcus canis sp. nov., a skin bacterium associated with infections in dogs. International journal of systematic and evolutionary microbiology 67(3):621-626.
Callon, C., C. Arliguie, and M. C. Montel. 2016. Control of Shigatoxin-producing Escherichia coli in cheese by dairy bacterial strains. Food microbiology 53(Pt B):63-70.
Collins, M. and D. Jones. 1981. Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implication. Microbiological reviews 45(2):316.
Desmazeaud, M. and J. Hermier. 1968. Isolement, purification et propriétés d’une protéase exocellulaire de Micrococcus caseolyticus. Pages 565-577 in Proc. Annales de Biologie Animale Biochimie Biophysique. EDP Sciences.
Endl, J., H. Seidl, F. Fiedler, and K. Schleider. 1983. Chemical composition and structure of cell wall teichoic acids of staphylococci. Archives of microbiology 135(3):215-223.
Evans, A. C. 1916. The bacteria of milk freshly drawn from normal udders. The Journal of Infectious Diseases:437-476.
Falk, D. and S. Guering. 1983. Differentiation of Staphylococcus and Micrococcus spp. with the Taxo A bacitracin disk. Journal of clinical microbiology 18(3):719-721.
Faller, A. and K.-H. Schleifer. 1981. Modified oxidase and benzidine tests for separation of staphylococci from micrococci. Journal of Clinical Microbiology 13(6):1031-1035.
Faller, A. H., F. Götz, and K.-H. Schleifer. 1980. Cytochrome-patterns of staphylococci and micrococci and their taxonomic implications. Zentralblatt für Bakteriologie: I. Abt. Originale C: Allgemeine, angewandte und ökologische Mikrobiologie 1(1):26-39.
Ghuysen, J. M., E. Bricas, M. Lache, and M. Leyh-Bouille. 1968. Structure of the cell walls of Micrococcus lysodeikticus. III. Isolation of a new peptide dimer, Nα-[L-alanyl-γ-(α-D-glutamylglycine)]-L-lysyl-D-alanine. Biochemistry 7(4):1450-1460.
Götz, F., T. Bannerman, and K.-H. Schleifer. 2006. The Genera Staphylococcus and Macrococcus. Pages 5-75 in The Prokaryotes: Volume 4: Bacteria: Firmicutes, Cyanobacteria. M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt, ed. Springer US, New York, NY.
Götz, F., E. Nürnberger, and K. Schleifer. 1979. Distribution of class‐I and class‐II D‐fructose 1, 6‐biphosphate aldolases in various staphylococci, peptococci and micrococci. FEMS Microbiology Letters 5(4):253-257.
Hargrove, R. E. and F. E. Mcdonough. 1964. Process of making low-fat ripened skim milk cheese. Google Patents.
Hill, L. 1959. The Adansonian classification of the staphylococci. Microbiology 20(2):277-283.
Hiramatsu, K., Y. Katayama, M. Matsuo, T. Sasaki, Y. Morimoto, A. Sekiguchi, and T. Baba. 2014. Multi-drug-resistant Staphylococcus aureus and future chemotherapy. Journal of Infection and Chemotherapy 20(10):593-601.
Jeffries, L., M. Cawthorne, M. HARRIS, B. COOK, and A. Diplock. 1968. Menaquinone determination in the taxonomy of Micrococcaceae. Microbiology 54(3):365-380.
Kaban, G. and M. Kaya. 2008. Identification of Lactic Acid Bacteria and Gram‐Positive Catalase‐Positive Cocci Isolated from Naturally Fermented Sausage (Sucuk). Journal of food science 73(8).
Kasik, R. L. and A. J. Luksas. 1971. Preparation of an elastic cheese product. Google Patents.
Kilpper, R., U. Buhl, and K. H. Schleifer. 1980. Nucleic acid homology studies between Peptococcus saccharolyticus and various anaerobic and facultative anaerobic Gram‐positive cocci. FEMS Microbiology Letters 8(4):205-210.
Kitching, J. S. and L. N. Farrell. 1936. STAPHYLOCOCCAL IMMUNITY12. American Journal of Epidemiology 24(2):268-284.
Kloos, W. E., D. N. Ballard, C. G. George, J. A. Webster, R. J. Hubner, W. Ludwig, K. H. Schleifer, F. Fiedler, and K. Schubert. 1998a. Delimiting the genus Staphylococcus through description of Macrococcus caseolyticus gen. nov., comb. nov. and Macrococcus equipercicus sp. nov., and Macrococcus bovicus sp. no. and Macrococcus carouselicus sp. nov. International journal of systematic bacteriology 48 Pt 3:859-877.
Kloos, W. E., C. G. George, J. S. Olgiate, L. Van Pelt, M. L. McKinnon, B. L. Zimmer, E. Muller, M. P. Weinstein, and S. Mirrett. 1998b. Staphylococcus hominis subsp. novobiosepticus subsp. nov., a novel trehalose-and N-acetyl-D-glucosamine-negative, novobiocin-and multiple-antibiotic-resistant subspecies isolated from human blood cultures. International journal of systematic and evolutionary microbiology 48(3):799-812.
Kwok, A. Y. and A. W. Chow. 2003. Phylogenetic study of Staphylococcus and Macrococcus species based on partial hsp60 gene sequences. International journal of systematic and evolutionary microbiology 53(1):87-92.
Lory, S. 2014. The Family Staphylococcaceae. Pages 363-366 in The Prokaryotes: Firmicutes and Tenericutes. E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson, ed. Springer Berlin Heidelberg, Berlin, Heidelberg.
LUDWIG, W., K.-H. SCHLEIFER, G. E. FOX, E. SEEWALDT, and E. STACKEBRANDT. 1981. A phylogenetic analysis of staphylococci, Peptococcus saccharolyticus and Micrococcus mucilaginosus. Microbiology 125(2):357-366.
Mannerová, S., R. Pantůček, J. Doškař, P. Švec, C. Snauwaert, M. Vancanneyt, J. Swings, and I. Sedláček. 2003. Macrococcus brunensis sp. nov., Macrococcus hajekii sp. nov. and Macrococcus lamae sp. nov., from the skin of llamas. International journal of systematic and evolutionary microbiology 53(5):1647-1654.
Martín, B., M. Garriga, M. Hugas, S. Bover-Cid, M. Veciana-Nogués, and T. Aymerich. 2006. Molecular, technological and safety characterization of Gram-positive catalase-positive cocci from slightly fermented sausages. International Journal of Food Microbiology 107(2):148-158.
Moreno, V. and F. Kosikowski. 1973. Degradation of β-casein by micrococcal cell-free preparations. Journal of Dairy Science 56(1):33-38.
Nahaie, M., M. Goodfellow, D. Minnikin, and V. Hajek. 1984. Polar lipid and isoprenoid quinone composition in the classification of Staphylococcus. Microbiology 130(9):2427-2437.
Ogston, A. 1882. Micrococcus Poisoning. Journal of Anatomy and Physiology 17(Pt 1):24-58.
Paul, F., F. Duchiron, and P. Monsan. 1990. Enzyme, its method of production and its application to the preparation of methyl N-(L-aspartyl-1) L-phenylalaninate. Google Patents.
Piard, J., M. El Soda, W. Alkhalaf, M. Rousseau, M. Desmazeaud, L. Vassal, and J. Gripon. 1986. Acceleration of cheese ripening with liposome-entrapped proteinase. Biotechnology letters 8(4):241-246.
Rosenbach, A. J. F. 1884. Mikroorganismen bei den Wundinfektionskrankheiten des Menschen. Bergmann.
Ruhland, G. and F. Fiedler. 1990. Occurrence and structure of lipoteichoic acids in the genus Staphylococcus. Archives of microbiology 154(4):375-379.
Schleifer, K.-H. 2015. Macrococcus. in Bergey’s Manual of Systematics of Archaea and Bacteria. John Wiley & Sons, Ltd.
Schleifer, K. 1973. Chemical composition of staphylococcal cell walls. Contributions to microbiology and immunology 1:13.
Schleifer, K., R. Kilpper-Balz, U. Fischer, A. Faller, and J. Endl. 1982a. Identification of “Micrococcus candidus” ATCC 14852 as a Strain of Staphylococcus epidermidis and of “Micrococcus caseolyticus” ATCC 13548 and Micrococcus varians ATCC 29750 as Members of a New Species, Staphylococcus caseolyticus. Vol. 32.
Schleifer, K. H. and O. Kandler. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriological Reviews 36(4):407-477.
SCHLEIFER, K. H., R. KILPPER-BÄLZ, U. FISCHER, A. FALLER, and J. ENDL. 1982b. Identification of “Micrococcus candidus” ATCC 14852 as a strain of Staphylococcus epidermidis and of “Micrococcus caseolyticus” ATCC 13548 and Micrococcus varians ATCC 29750 as Members of a New Species, Staphylococcus caseolyticus. International journal of systematic and evolutionary microbiology 32(1):15-20.
Schleifer, K. H. and W. E. Kloos. 1975. A simple test system for the separation of staphylococci from micrococci. Journal of Clinical Microbiology 1(3):337-338.
Schwendener, S., K. Cotting, and V. Perreten. 2017. Novel methicillin resistance gene mecD in clinical Macrococcus caseolyticus strains from bovine and canine sources. Scientific reports 7:43797.
Silvestri, L. G. and L. R. Hill. 1965. Agreement Between Deoxyribonucleic Acid Base Composition and Taxometric Classification of Gram-Positive Cocci. Journal of Bacteriology 90(1):136-140.
Vos, P., G. Garrity, D. Jones, N. R. Krieg, W. Ludwig, F. A. Rainey, K.-H. Schleifer, and W. B. Whitman. 2011. Bergey’s manual of systematic bacteriology: Volume 3: The Firmicutes. Vol. 3. Springer Science & Business Media.
Wu, Y., C. Cui, W. Sun, B. Yang, and M. Zhao. 2009. Effects of Staphylococcus condimenti and Micrococcus caseolyticus on the volatile compounds of Cantonese sausage. Journal of food process engineering 32(6):844-854.
Yoovidhya, T., D. Combes, and P. Monsan. 1986. Kinetic and thermal stability studies of rulactine, a proteolytic enzyme fromMicrococcus caseolyticus. Biotechnology letters 8(5):333-338.
Cite This Work
To export a reference to this article please select a referencing stye below:
Related Services
View allRelated Content
All TagsContent relating to: "Biology"
Biology is the scientific study of the natural processes of living organisms or life in all its forms. including origin, growth, reproduction, structure, and behaviour and encompasses numerous fields such as botany, zoology, mycology, and microbiology.
Related Articles
DMCA / Removal Request
If you are the original writer of this dissertation and no longer wish to have your work published on the UKDiss.com website then please:




