Protein Kinase Cell Signalling and Inhibition
Info: 4975 words (20 pages) Dissertation
Published: 10th Jun 2021
Tagged: Genomics
Protein Kinases: potential drug targets for altering signal transduction.
Introduction
It is estimated that protein kinases are responsible for making up around 2% of the human genome with 500 protein kinase genes (Subramani et al., 2013). Protein kinases play a crucial role towards supporting cell health through their regulatory properties. These kinases are a superfamily of enzymes which are responsible for conducting a reaction known as phosphorylation. Kinases have the ability to regulate the biological activity of proteins through this catalytic activity which will be discussed later on throughout the dissertation.
The objective of this research topic is to explore many areas of protein kinases, limited to their classification, structure, function in cell signalling. The remainder of this dissertation will focus on a chosen protein kinase, investigating its role in signalling, how they can be inhibited and the potential drugs which can be applied to target its inhibition.
Types of protein kinases
Protein kinases are essential for regulating and coordinating functions of cell growth, cell motility, cell proliferation and differentiation, to name a few. It is believed that anywhere from 30-50% of proteins within the human genome can undergo phosphorylation as mediated by these protein kinases (Subramani et al., 2013). Early studies have argued that protein kinases may be classified according to the amino acid residues that are phosphorylated as per the Nomenclature Committee of the International Union of Biochemists. Protein kinases are classified into 5 groups under the EC 2.7 subtype of transferases, a phosphorous-containing group (International Union of Biochemistry and Molecular Biology, 2021). Within this transferases subtype are the 5 types of protein kinases as follows (Hunter, 1991):
- Protein-serine/threonine kinases
- Protein-tyrosine kinases
- Protein-histidine kinases
- Protein-aspartyl/glutamyl kinases
- Protein-cysteine
The 5 major types of protein kinases are phosphotransferases where they have the capability to transfer a phosphate group onto an amino acid acceptor. According to Hunter, it is most logical to categorise protein kinases based on the acceptor amino acid instead of their specific substrate (Hunter, 1991).
Role in cell signalling
The maintenance of the intracellular transduction system is of paramount importance to regulate cell proliferation and differentiation. Protein kinases act as biological safety parameters which manages the signalling pathways by turning on and off effector molecules . These are essentially enzymes which trigger the phosphorylation of the protein substrate (Cormier and Woodgett, 2016). Protein kinases are responsible for regulatory events known as phosphorylation which is a post translational modification where the function of a protein is modulated (Bononi et al., 2011). Phosphorylation causes modifications of specific residues in a number of ways. This includes altering the structure of residues through amending the way in which the protein folds, altering their substrate affinity and stability (Bononi et al., 2011). As previously mentioned within the classifications of proteins, phosphorylation occurs through the addition of a phosphate which is transferred from the protein kinase to the amino acid in a protein substrate. A schematic of the phosphorylation reaction can be visualised in figure 1.

Figure 1 - Reaction mechanism of phosphorylation as mediated by protein kinases. A phosphate group is donated from adenosine triphosphate (ATP), the energy currency. ATP is reduced to adenine diphosphate (ADP) to form a phosphorylated protein, a protein which goes under conformational change upon addition of phosphate group. This conformational change has the ability to either activate or deactivate the protein depending on the type of regulation needed (Weber, 2010).
This reaction mechanism is achieved through the addition of a phosphate group (figure 1). Kinases induce phosphorylation using ATP as a phosphate source or also known as a phosphate donor. This phosphate group is accepted by an amino acid in a substrate protein (Hunter, 1991). Phosphate groups allow for conformational changes to the protein, thereby activating or inactivating the protein. This method of turning proteins on and off per se, is some of the natural parameters within the cell that can prevent disease such as cancer caused by aberrant cells. Since kinases are linked to such diseases, researchers can design drugs to target kinases and in turn, inhibit their activity.
Kinases encoded by the human genome
Kinases encodes by the human genome are coined as the kinome. A kinome is a subset of an organism’s genome which consists of all kinases expressed within the cell. Within the human kinome both lipid and protein kinases are present. In the case of protein kinases, it is made up of two domains, the eukaryotic protein kinase and atypical protein kinase domain (figure 2).
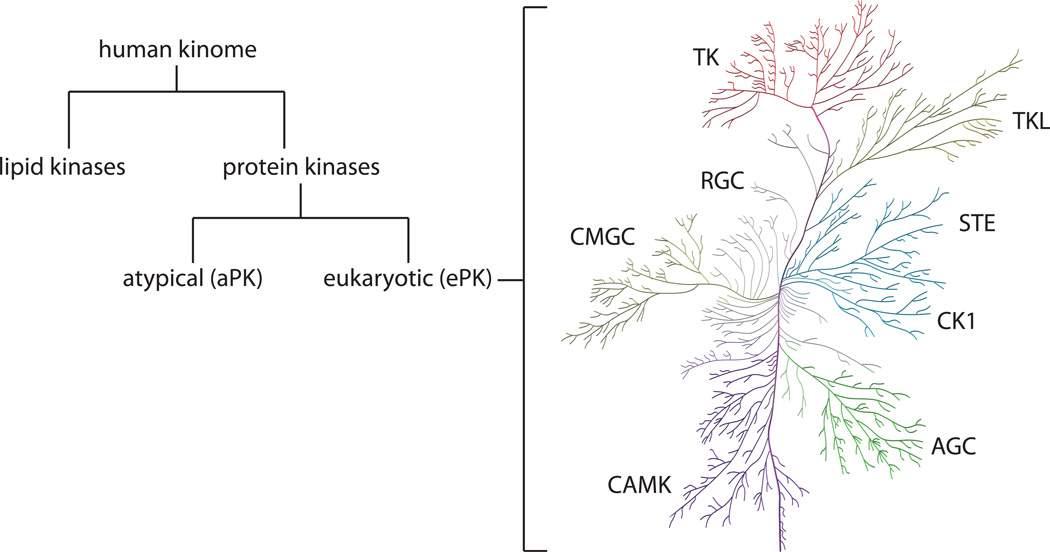
Figure 2 - Classification of the human kinome into lipid kinases and protein kinases. Protein kinases are further classified into two domains being eukaryotic and atypical protein kinases. These eukaryotic protein kinases can be categorised even further as seen on the right-hand side in the kinome tree.
Eukaryotic protein kinases can be categories even further into 9 groups based on sequence similarity as per table 1.
|
Protein Kinase |
Description |
Kinase members & pathways |
|
TK |
Tyrosine Kinase consisting of receptor tyrosine kinase (RTK) and non-receptor tyrosine kinase |
Growth factors e.g., human epidermal growth factor and insulin receptor |
|
TKL |
Resembles TK sequence and for that reason is termed tyrosine kinase-like. Contains both receptor and non-receptor protein kinases |
RAF (Rapidly accelerated fibrosarcoma) and transforming growth factor beta receptors (TGFβ) |
|
STE |
Homologs of yeast proteins STE20/11/7 |
Tumour suppressor p21-activated kinases (Paks) |
|
CK1 |
Casein kinase 1 family, containing Serine/Threonine selective enzymes |
Regulates transcription and cell division, linked to the degradation and monitoring of p53 tumour suppressor |
|
AGC |
Protein Kinase A (PKA), G (PKG) & C (PKC) families which are related to serine/threonine kinases |
PKA relates to cAMP-dependent kinase 1, PKG relates cGMP-dependent kinase 1 and PKC is protein kinase C all of which are associated to cell proliferation |
|
CAMK |
Calcium or calmodulin-dependent protein kinase |
Cell checkpoint kinases that signal a stop in cell cycle and lead to cell repair. CHK 1 and CHK2 are enzymes within this family which are safety parameters in the cell cycle |
|
CMGC |
Named after families such as cyclin-dependent kinases (CDK) and mitogen-activated protein kinases (MAPK) |
These are a family of kinases which are central to cell cycle regulation which mediate processes such as cell proliferation and cell death (apoptosis) |
|
RGC |
Receptor guanylyl cyclase (RGC) |
Responsible for the conversion of GTP to cyclic GMP. Family consisting of just pseudokinases |
|
Others |
Low sequence similarity to major classifications of PKs |
Casein kinase 2 (CK2) which plays a key role in cell differentiation |
Table 1: Major classifications of eukaryotic protein kinases which are based on sequence similarity. A brief description of each family is provided alongside the associated pathways in cell signalling. Some examples of kinases within these classifications are given with their role in the relevant pathway (Duong‐Ly and Peterson, 2013; Cell Signalling Technology, 2021)
As per table 1, there are 9 major classifications of eukaryotic protein kinases (ePKs) listed including eukaryotic typical PKs such as TK, TKL, STE, AGC, CK1, CAMK, GMGC, RGC and others which lack sequence similarity to the 8 others. Atypical protein kinases (aPKs) are another primary type of protein kinase, these generally lack sequence similarity. Despite this, it is possible for the aPKs to mimic the typical confirmational fold of ePKs. Some of the most notable families within the aPKs include the breakpoint cluster region gene (BCR), pyruvate dehydrogenase kinase and mTOR (mammalian target of rapamycin). BCR and mTOR both display serine/kinase activity (NCBI, 2021) where BCR displays regulatory functions towards guanine triphosphate (GTP) binding proteins (Uniprot, 2021). In the case of mTOR, it has a functional role in multiple pathways linked to cell proliferation and growth (Duong-Ly and Peterson, 2013).
Structure of Protein Kinases
In general, all eukaryotic protein kinase (ePK) domains follow a similar structure. It is estimated that the length of an ePK domain is around 250 amino acids long. This domain consists of two lobes, a cleft or hinge region, a phosphate binding loop (P-loop) and an activation loop (A-loop) (Duong-Ly and Peterson, 2013).
The first lobe is found at the N-terminal consisting of predominantly beta structures with a standalone alpha helix structure termed as helix C. The other lobe is the C-terminal which is much larger in size and in this case, alpha helical structures are mostly present. These lobes each have a functional role for catalytic activity with important structural motifs (Kobe and Kemp, 2003). The c-lobe contains residues including a glycine-rich motif which interacts with the phosphate acceptor, forming a P-loop that anchors the phosphate groups donated from ATP. Another important motif is the A-loop which contains phosphorylation sites providing a means for the substrate binding (Kobe and Kemp, 2010). These lobes are separated through a short linker hinge or cleft with a relatively flexible backbone.
More specifically, the structure of the mitogen-activated protein kinase or MAPK will be analysed and described in more detail via the available bioinformatics tools.
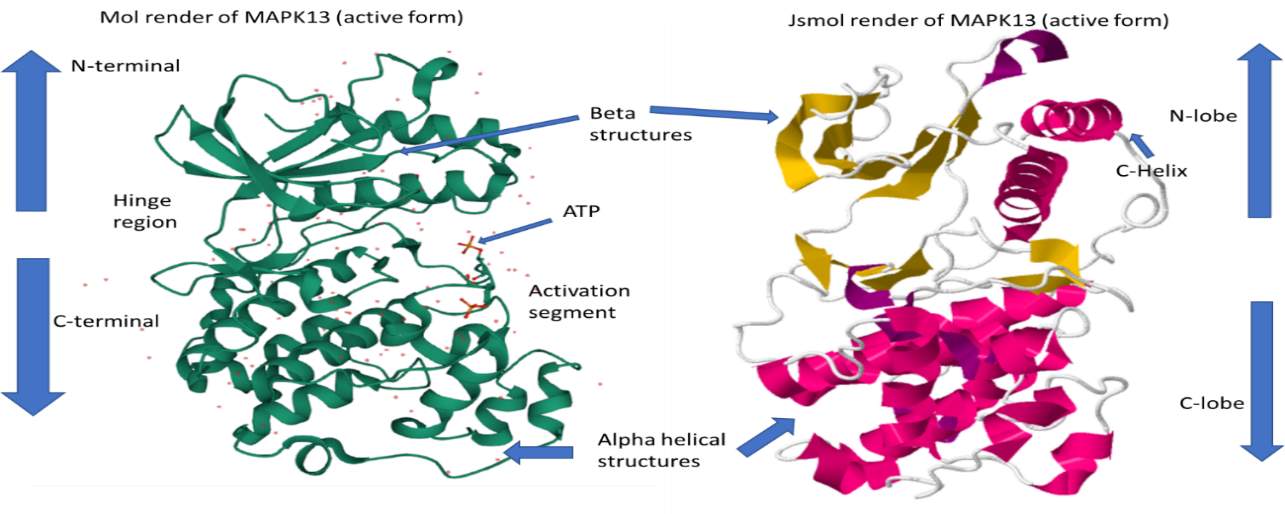
Figure 3 – 3D render of protein kinase MAPK13 in its active form, PDB ID: 4MYG. Structures gathered from the protein data bank (RSCB.org) and labelled with the standard protein kinase domains. On the left-hand side is a 3D model of MAPK13 rendered from the Mol tool (Sehnal et al., 2018). On the right-hand side is the MAPK13 protein kinase rendered in Jsmol bioinformatics tool. MAPK13 has distinct features of protein kinase, including a C-lobe, N-lobe, a hinge region, activation region and an ATP pocket.
The structure of MAPK13 (active form) is displayed in figure 3. Protein Kinase MAPK13 (fig. 3) is in its active form where it has been phosphorylated to turn on the kinase. This is evident due to the presence of ATP molecules in the activation segment of the structure. MAPK13 is a eukaryotic protein and subsequently shares the same basic structure of containing two lobes, a C-lobe with alpha helices and a N-lobe with mainly beta structures and one important helical structure (c-helix). Other domains include the hinge region with joins the N and C-terminal and an ATP pocket or activation segment.
Mitogen-activated protein kinases (MAPK)
As mentioned in the introduction, the remainder of the dissertation will focus on a chosen kinase. The author has decided to investigate into MAPK enzymes; how it is involved in the signalling pathway and how one may identify potential inhibitors and drug targets. Mitogen- protein kinases relay extracellular signals to produce intracellular responses, playing a key role in activated regulating cellular activities. MAPK phylogenetically belong to the CMGC kinase family which also contains cyclic-dependent kinases (table 1). MAPK kinases target substrates on serine/threonine residues. These enzymes act on serine and threonine amino acids by phosphorylating the OH group found in the residues. MAPK kinases are switched on through a wide range of stimuli together with growth factors, cytokines, and cell adherence. In eukaryotic cells, there are an abundant of MAPK pathways which regulate cell apoptosis, motility, metabolism, and gene expression (Cargnello and Roux, 2011). Mammals have 14 types of kinases classified into the MAPK family; they are broadly categorised into either conventional or atypical MAPKs (figure 4).
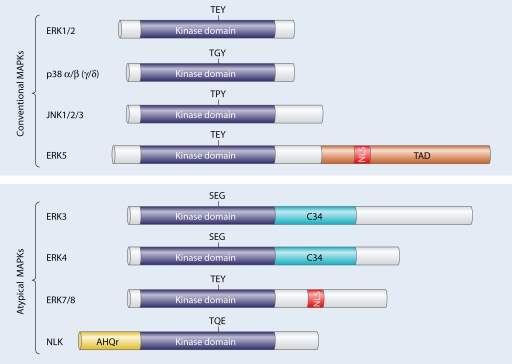
Figure 4 – Family tree of mammalian MAPK kinases. An overview of conventional and atypical MAPKs are provided. Each MAPK kinase have serine/threonine kinase domains and some with additional domains such as TAD (transactivation domain), C34 (region which is conversed in ERK3/4 kinases) and AHQr (domain rich in amino acids Alanine, Histidine and Glutamic acid). Another notable domain is the nuclear localisation sequence (NLS) which is found in the ERK5 and ERK7/8 protein kinase (Cargnello and Roux, 2011).
As previously mentioned, there are quite a number of protein kinases which are associated with the MAPK family (fig. 4). Conventional MAPKS are comprised of the following regulatory kinases:
- ERK 1/2 – Extracellular signal-regulated kinases 1 and 2
- JNK1/2/3 –c-JUN N-terminal kinase
- p38 – MAPK with isoforms p38 (α, β, γ and δ)
- ERK5 – Extracellular signal-regulated kinase 5
Atypical MPAKs are as follows:
- ERK 3, 4, 7/8
- NLK - Nuclear localisation sequence
In the following sections, an overview of a few kinase will be provided alongside with inhibitors and potential drug targets. Since JNK, P38 and ERK1/2 kinases have been extensively researched, it is rather appropriate to focus on these kinases specifically.
ERK 1/2 pathway and inhibition
The mammalian ERK 1/2 module consists of MAPK kinase kinase (MAPKKKs), rapidly accelerated fibrosarcoma A and B (Raf), mitogen-activated protein kinase kinase (MEK/MAP2K) and ERK 1 and 2 (Cargnello and Roux, 2011). The ERK 1/2 module within latent cells is located in the cytoplasm and is translocated to the nucleus upon receiving an extracellular signal (Cargnello and Roux, 2011).
The EKR1/2 module is activated by cell surface receptors such epidermal growth factor and nerve growth factor. Upon the binding of a growth factor or hormone to the receptor, rat sarcoma (Ras) is activated causing a cascade of catalytic reactions which eventually leads to the activation the ERK 1/2 kinases (figure 5). ERK 1/2 acts on tyrosine residues during phosphorylation. Once phosphorylated, the tyrosine residues are transformed into a specific binding site for proteins consisting of phosphotyrosine-binding domains.
ERK1/2 is one involved in one of the few MAPKs signalling pathways. The route of this signal transduction pathway follows a specific order as per figure 5.
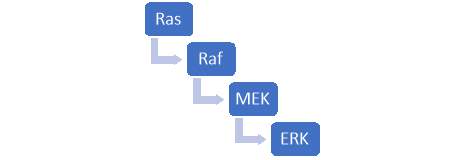
Figure 5 – MAPK pathway which begins with the activation of Ras. A ligand such as a growth factor attaches itself to a tyrosine receptor and in turn activates Ras proteins. The Ras proteins then bind to Guanosine diphosphate to form guanosine triphosphate and cause a confirmational change in the Ras protein. As a result, a cascade reaction occurs as seen above. Ras induces Raf and in turn Raf activates MEK which activates ERK.
MEK is responsible for the activation or phosphorylation of ERK, so far it is the only known kinase which has the capability to turn on ERK 1/2. Once activated, ERK 1/2 eventually leads to cell proliferation, survival, and differentiation. Dysregulation within this pathway can lead to hyperactivity of ERK 1/2 leading to the potential of oncogenesis. Since this is a potential risk, targeting the ERK 1/2 module is of great interest for the prevention of cancer and can be studied for developing drugs for targeted therapy. Like all pathways, ERK 1/2 can be inhibited in two possible ways, through indirect and direction inhibition with either an ATP-competitive or non-ATP-competitive inhibitor.
ERK 1/2 can be directly or indirectly inhibited. The mode of indirect inhibition involves blocking the activation of a specific protein within the cascade reaction, the specific protein being a kinase which leads to the activation of the protein of interest. For example, within the MAPK pathway, one could block Ras, Raf or MEK as demonstrated in figure 5. Once a single step within the cascade reaction is blocked, it prevents the activation of the target protein (ERK 1/2). Direct inhibition, on the other hand, can target the protein kinase domain to prevent binding to its specific substrate an in turn, block phosphorylation.
A few pharmaceutical companies have invested into the use of MEK 1/2 for indirectly inhibiting ERK proteins. It is a non-ATP-competitive inhibition since they do not interact with active phosphorylated kinases but rather demonstrates a strong interaction with the unphosphorylated species (Cargnello and Roux, 2011). Two classes of MEK inhibitors were developed in 1990s, PD98059 and U0126 which specifically target MEK. Both of these are considered as non-ATP-competitive which means they rely on blocking specific protein-protein interactions such as blocking the binding domain and the target protein from interacting. One limitation outlined by Cargnello and Roux was that although the therapeutic target is effective, it is still quite low in bioavailability. A successful application of this drug would help to put a stop to the development of oncogenesis. Mutations within Ras and Raf would disrupt the maintenance of cell cycle and leading to ERK 1/2 activation causing proliferation of aberrant cells (Cargnello and Roux, 2011).
p38 pathway and inhibition
p38 is a member within the MAPK family and has been mentioned in slight detail previously in the dissertation. As aforementioned, the p38 module contains four isoforms, p38α, β, γ and δ. These kinases are sensitive to environmental stress such as UV irradiation, oxidative stress, and inflammatory cytokines such as interleukin-1 (Cargnello and Roux, 2011). Similar to what was seen within the ERK module, p38 isoforms are activated through a cascade of reactions (figure 6). The p38 pathway starts off with extracellular signal followed by the activation of MAPKKK, turning on MAPKK such as MKK 3/4/6, the main protein kinases associated with the activation of p38 and its isoforms (Cargnello and Roux, 2011). To aid in the visualisation of this reaction, a step-down process chart is provided as per figure 6.

Figure 6 – Cascade of reactions leading to the activation of p38 isoforms. An extracellular signal activates MAPKKK leading to the phosphorylation of MKK3/4/6 which are responsible for turning on p38. Information gathered from Cargnello and Roux (2011).
The protein kinases MKK3/6 are considered to be the main protein kinases which activate p38 and its isoforms. MKK 4 is one of the lesser-known kinases for p38 since its research is quite restricted and also shows a low level of activity towards p38 phosphorylation. MKK6 has the capability to phosphorylate all of the p38 isoforms, while on the other hand, MKK3 can only activate a select few isoforms. MKK3 has more of an affinity to α, δ and γ isoforms since it specifically phosphorylates these p38 variants.
Upon activation of either MKK3/4/6, p38 contributes to the proliferation of cells and their survival. Studies have also shown that the p38 module plays a vital role in the immune and inflammatory response since its activated by extracellular stimuli which mediate inflammatory responses, these include cytokines and chemokines (Cargnello and Roux, 2011). These p38 work towards the regulation of cytokines by controlling their expression, this is achieved through the modification of transcription factors and their stability.
Due to the fact that p38 can induce inflammatory responses, there has been therapeutic targets developed to mimic the functions of p38 isoforms. The anti-inflammatory drugs SB203580 and SB202190 can act as inhibitors of p38α and p38β by preventing ATP-binding (Cargnello and Roux, 2011). This is an ATP-competitive mode of inhibition where since the therapeutic agent competes with the isoforms in the ATP binding. A more potent alternative to the aforementioned inhibitors is the drug BIRB0796, which indirectly competes with ATP to prevent the activation of p38α and p38β isoforms. One of the most obvious limitations associated with p38 inhibition is that the targets are specific to p38α and p38β with none block the phosphorylation of γ and δ isoforms.
JNK pathway and inhibition
The JNK module is quite similar to p38 since it is activated through cellular stress. As a result, this kinase can also be termed as the stress-activated protein kinase (SAPK). Some of the cellular stress include UV irradiation, DNA-damaging agents or mutagens and oxidative stress (Cargnello and Roux, 2011). The JNK pathway, much like the p38 pathway, is activated by an extracellular stimulus with leads to the activation of MAPKKK leading to a cascade of reactions. A series of reactions takes place as follows:

Figure 7 – Cascade of reactions leading to the activation of JNK and its isoforms. Data compiled from Cargnello and Roux (2011).
The JNK module contains three isoforms (figure 7) including JNK 1, 2 and 3 which can alternatively be named SAPKγ, β and α. Isoforms JNK1 and JNK2 play a crucial role in cell proliferation regulation. These isoforms have found to positively regulate the transcription factor c-Jun which transduces mitogen responses leading to cell growth. This is of great importance within cancer research since c-Jun is one of the most researched proteins in the cell signalling pathway. It is a proto-oncogene with a potential to cause normal cells to turn carcinogenic. JNK is also known to regulate several other transcription factors such as p53 (tumour suppressor) and Elk-1 (Cargnello and Roux, 2011).
At current, there are two main JNK inhibitors which have become available within the past few decades including SP600125 (JNK inhibitor II) and AS601245 (JNK inhibitor V) (Cargnello and Roux, 2011). These are reversible, ATP-competitive inhibitors which are effective across all JNK isoforms. The inhibitors induce a change in the conformational shape of the target enzyme which hinders kinase activity. These are important targets for preventing the dysregulation of the c-Jun gene which is proto-oncogenic in nature. However, there are some pitfalls associated with the drugs outlined above. Literature has demonstrated that JNK inhibitor II and V lack selectivity upon contact with purified protein kinases raising a question about the efficacy and reliability of the therapeutic agents.
Conclusion
An overview of kinases was provided throughout the dissertation including their classification, the protein kinases encoded by the human genome, their structure and role in signalling. Protein kinases can be classified into the amino acid substrates which they act on and in the case of eukaryotic PKs, they can be categorised broadly into atypical and typical families. More specifically, an account of typical protein kinases was provided including their main classifications TK, TKL, STE, CK1, AGC, CAMK, CMGC, RGC and others. These protein kinases contribute to cell signalling through phosphorylation, a form of post-translational modifications. The structure of eukaryotic protein kinases follow a similar structure with distinctive domains including the activation loop, a c-lobe and n-lobe and hinge region which can be seen in figure 3. Once an overview of protein kinases was provided, the author narrowed down the scope to a specific protein kinase, MAPK. This a type of eukaryotic protein kinase which is affiliated with the CMGC family. A review of the MAPK signalling pathways was provided with the kinase subfamilies. ERK1/2, JNK and p38 are the most extensively researched MAPKS and so were investigated for their associated pathways and potential inhibitors. The inhibitors outlined were scrutinised to detect their limitations as drug targets. Overall, protein kinases and their role in cell signalling can be the key towards developing drug targets for the prevention of oncogenesis.
References
Bononi, A., Agnoletto, C., De Marchi, E., Marchi, S., Patergnani, S., Bonora, M., Giorgi, C., Missiroli, S., Poletti, F., Rimessi, A. and Pinton, P. (2011). Protein Kinases and Phosphatases in the Control of Cell Fate. Enzyme Research, [online] 2011, pp.1–26. Available at: https://www.hindawi.com/journals/er/2011/329098/ [Accessed 22 Feb. 2021].
Cargnello, M. and Roux, P.P. (2011). Activation and Function of the MAPKs and Their Substrates, the MAPK-Activated Protein Kinases. Microbiology and Molecular Biology Reviews, [online] 75(1), pp.50–83. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3063353/#r396 [Accessed 25 Feb. 2021].
Cell Signalling Technology. (2021). Human Protein Kinases Overview | Cell Signalling Technology. [online] Available at: https://www.cellsignal.com/learn-and-support/protein-kinases/human-protein-kinases-overview [Accessed 16 Feb. 2021].
Cormier, K.W. and Woodgett, J.R. (2016). Protein Kinases: Physiological Roles in Cell Signalling. eLS, [online] pp.1–9. Available at: https://onlinelibrary.wiley.com/doi/abs/10.1002/9780470015902.a0002710.pub3#:~:text=Protein%20kinases%20are%20enzymes%20that,key%20feature%20of%20cellular%20signalling.&text=Crosstalk%20with%20other%20post%E2%80%90translational,in%20nearly%20every%20physiological%20process. [Accessed 22 Feb. 2021].
Duong‐Ly, K.C. and Peterson, J.R. (2013). The Human Kinome and Kinase Inhibition. Current Protocols in Pharmacology, [online] 60(1). Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4128285/ [Accessed 16 Feb. 2021].
Hunter, T. (1991). [1] Protein kinase classification. Methods in Enzymology, [online] pp.3–37. Available at: https://www.sciencedirect.com/science/article/pii/007668799100125G [Accessed 15 Feb. 2021].
International Union of Biochemistry and Molecular Biology (2011). Enzyme Nomenclature. [online] Nomenclature Committee of the International Union of Biochemistry and Molecular Biology. Available at: https://www.qmul.ac.uk/sbcs/iubmb/enzyme/ [Accessed 15 Feb. 2021].
Kobe, B. and Kemp, B.E. (2003). Principles of Kinase Regulation. Handbook of Cell Signalling, [online] pp.539–542. Available at: https://www.sciencedirect.com/science/article/pii/B9780121245467504502 [Accessed 23 Feb. 2021].
NCBI (2021). BCR BCR activator of RhoGEF and GTPase [Homo sapiens (human)] - Gene - NCBI. [online] NCBI. Available at: https://www.ncbi.nlm.nih.gov/gene/613 [Accessed 16 Feb. 2021].
Subramani, S., Jayapalan, S., Kalpana, R. and Natarajan, J. (2013). HomoKinase: A Curated Database of Human Protein Kinases. ISRN Computational Biology, [online] 2013, pp.1–5. Available at: https://www.hindawi.com/journals/isrn/2013/417634/ [Accessed 15 Feb. 2021].
Uniprot (2021). BCR - Breakpoint cluster region protein - Homo sapiens (Human) - BCR gene & protein. [online] Uniprot.org. Available at: https://www.uniprot.org/uniprot/P11274 [Accessed 16 Feb. 2021].
Weber, T.J. (2010). Protein Kinases. Comprehensive Toxicology, [online] pp.473–493. Available at: https://www.sciencedirect.com/science/article/pii/B9780080468846002256 [Accessed 15 Feb. 2021].
Cite This Work
To export a reference to this article please select a referencing stye below:
Related Services
View allRelated Content
All TagsContent relating to: "Genomics"
Genomics is an interdisciplinary science that focuses on the study of all genes of an organism, their functions and how they interact with each other and their environment. The complete DNA, including all genes, of an organism is a genome.
Related Articles
DMCA / Removal Request
If you are the original writer of this dissertation and no longer wish to have your work published on the UKDiss.com website then please:




